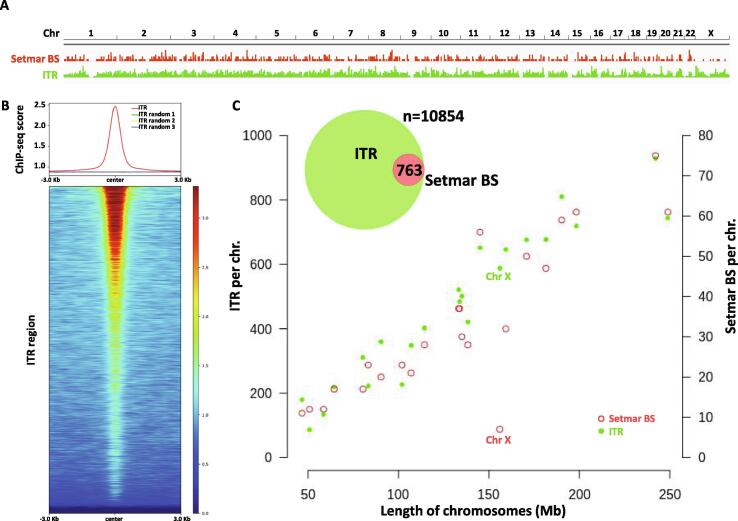
Fig. 5.
SETMAR-HA preferentially binds to inverted terminal repeat (ITR) sequences. (A) Genome browser track showing the distribution of SETMAR-HA ChIP peaks (blue) and ITR motifs (green) for each autosome and the sex chromosome (chrX). (B) The summit of SETMAR-HA signal perfectly coincides with ITRs (red curve and heatmap). The ChIP signal is depleted over random sites (yellow, green, and blue curves). (C) Upper panel: Proportional Venn diagram showing the overlap of SETMAR ChIP peaks and ITR motifs. Of the 764 SETMAR-HA peaks, 763 sites (99.9%) are associated with ITRs. Of the 10,854 ITRs, 1227 motifs (11.3%) are localized in SETMAR-HA peaks. Lower panel: Chromosomal distribution of SETMAR-HA binding sites and ITR motifs is strongly correlated. The number of SETMAR-HA peaks and ITRs were plotted as a function of chromosome length. They show significant covariation and correlation with chromosome length (Spearman r = 0.89; p < 0.001). Dots: chromosomes. Chromosome X represents an outlier in terms of ChIP peak numbers. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)