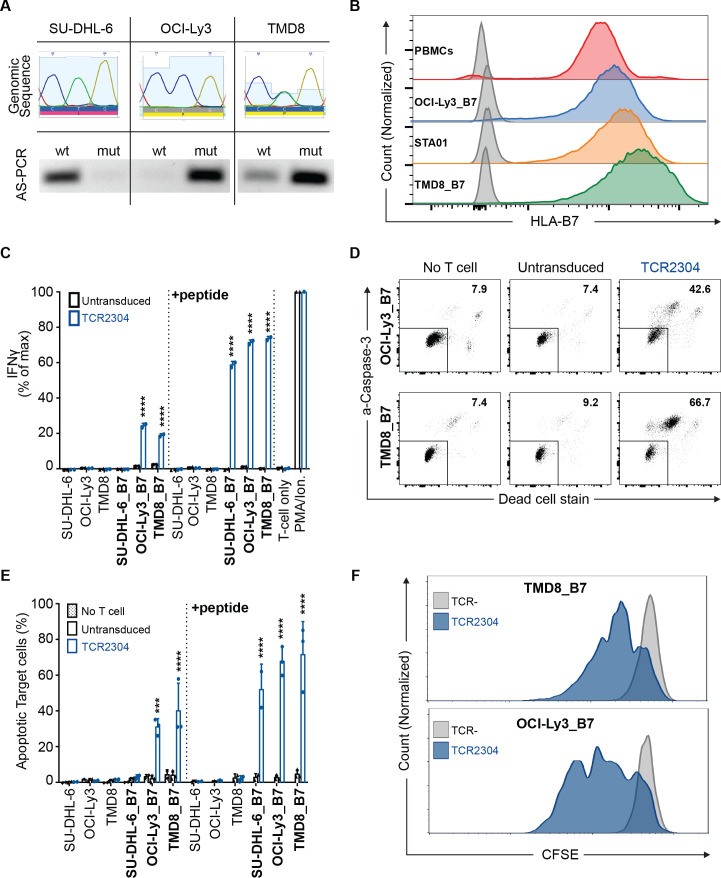
Figure 4.
Mutation-specific activity of TCR-T cells against DLBCL cell lines with intrinsic MyD88 L265P expression. (A) Genomic sequence and allele-specific (AS) PCR analysis of lymphoma cell lines: SU-DHL-6 (GBC-like DLBCL, WT MyD88), OCI-Ly3 (ABC-like DLBCL, homozygous MyD88 L265P) and TMD8 (ABC-like DLBCL, heterozygous MyD88 L265P). (B) Side-by-side comparison of HLA*B07:02 expression of virally transduced lines versus natural expression in PBMCs isolated from an HLA*B07:02-positive donor and an HLA*B07:02-positive B-LCL line (STA01) measured by flow cytometric staining. (C) Activation analysis of TCR2304-transduced T cells via IFN-γ ELISA, after 16 hours of coculture with lymphoma cells. Cell lines virally transduced to express HLA*B07:02 are shown as ‘cell line_B7’. Results from two different blood donors plotted with error bars showing SD significance analysis by two-way ANOVA: ****p<0.0001. (D) Representative flow cytometric viability analysis of lymphoma cells expressing HLA*B07:02 after 16 hours of coculture with T cells (as explained in figure 3). (E) Viability analysis of lymphoma cells with or without HLA*B07:02 expression. T cells of three different donors were used, error bars with SD significance analysis by two-way ANOVA: ***p<0.001, ****p<0.0001. (F) Antigen-induced proliferation of TCR2304-transduced T cells following 72 hours of coculture with HLA*B07:02-positive OCI-Ly3 and TMD8 cells. T cells were labeled with CFSE to trace proliferation prior to coculture. ANOVA, analysis of variance; DLBCL, diffuse large B-cell lymphoma; IFN-γ, interferon gamma; PBMC, peripheral blood mononuclear cell; TCR, T-cell receptor; WT, wild type.