Figure 2.
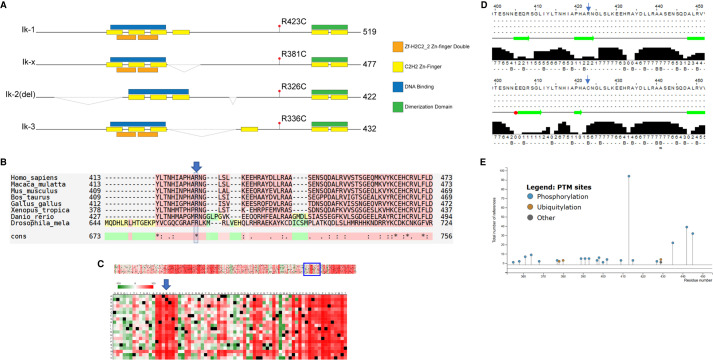
Protein sequence conservation and structural prediction suggests IKZF1 p.R381C is pathogenic. (A) The four functional isoforms of IKZF1 depicted with predicted Zn finger, DNA binding domains, and location of the variant p.R423C in isoform Ik-1. The variant location is p.R381C in isoform Ik-x. Isoform Ik-x is the isoform used in the described biochemical experiments in this manuscript. Note that the figure is not drawn to scale. (B) M-Coffee alignment reveals strict conservation of p.R381 (blue arrow and box) across orthologs from evolutionarily divergent species. (C) “OpenPredictProtein-effect of point mutation” algorithm predicts any changes at the p.R381C position (blue arrow) to be highly deleterious. (D) Jpred server prediction of secondary structure indicates that a R → C mutation (top vs. bottom panel, blue arrow heads) at p.R423 (equivalent to p.R381C in isoform Ik-x) would result in a loss of a B-sheet and potentially alter secondary structures up to 18 amino acid residues upstream (broad green arrows represent predicted B-sheets; red box indicates potential helical structure). (E) Phosphosite Plus analysis of IKZF1 Ik-1 isoform indicates post-translational modification sites near the p.R423 residue. Of note are p.S427-phos, p.K429-Ub, and p.K429-SUMO.