Abstract
Tissue function and homeostasis reflect the gene expression signature by which the combination of ubiquitous and tissue-specific genes contribute to the tissue maintenance and stimuli-responsive function. Enhancers are central to control this tissue-specific gene expression pattern. Here, we explore the correlation between the genomic location of enhancers and their role in tissue-specific gene expression. We find that enhancers showing tissue-specific activity are highly enriched in intronic regions and regulate the expression of genes involved in tissue-specific functions, whereas housekeeping genes are more often controlled by intergenic enhancers, common to many tissues. Notably, an intergenic-to-intronic active enhancers continuum is observed in the transition from developmental to adult stages: the most differentiated tissues present higher rates of intronic enhancers, whereas the lowest rates are observed in embryonic stem cells. Altogether, our results suggest that the genomic location of active enhancers is key for the tissue-specific control of gene expression.
Multiple layers of molecular and cellular events tightly control the level, time, and spatial distribution of expression of a particular gene. This wide range of mechanisms, known as gene regulation, defines tissue-specific gene expression signatures (Melé et al. 2015), which account for all the processes controlling the tissue function and maintenance, namely tissue homeostasis. Both the level and spatiotemporal pattern of expression of a gene are determined by a combination of regulatory elements (REs) controlling its transcriptional activation. Most genes contributing to tissue-specific expression signatures are actively transcribed in more than one tissue but at different levels and with distinct patterns of expression in time and space, suggesting that the regulation of these genes is different across tissues. Nevertheless, ∼10%–20% of all genes are ubiquitously expressed (housekeeping genes), and they are involved in basic cell maintenance functions (Eisenberg and Levanon 2013; Pervouchine et al. 2015; Zabidi et al. 2015).
cis-REs (CREs) are distributed across the whole genome, and their histone signature correlates with the transcriptional control they exert over their target genes (Hawkins et al. 2010; Choukrallah et al. 2015; Chen et al. 2019). The activation of CREs depends on several epigenetic features, including combinations of different transcription factors’ binding sites, and it is positively correlated with the H3K27ac histone modification signal (Heintzman et al. 2007; Heinz et al. 2015). Epigenetic features in specific tissues may change throughout the lifespan of individuals. During development, embryos undergo morphological and functional changes. These changes shape cell fate and identity as a result of tightly regulated transcriptional programs, which in turn are intimately associated with CREs’ activity and chromatin dynamics (Gilbert et al. 2003; Rand and Cedar 2003; Shlyueva et al. 2014; Bonev et al. 2017).
Many key CREs known to regulate gene expression have been reported to locate in introns of their target genes (Ott et al. 2009; Kawase et al. 2011). However, it is unknown whether this is either a sporadic feature associated with certain types of genes—for instance, long genes, such as HBB (also known as beta-globin) (Gillies et al. 1983) or CFTR (Ott et al. 2009)—or a common regulatory mechanism to most genes (Khandekar et al. 2007; Levine 2010) or a pattern of biological significance. To delve into this question, we analyzed the genomic location of CREs across a panel of 70 adult and embryonic human cell types available from the Encyclopedia of DNA Elements (ENCODE) Project (The ENCODE Project Consortium 2020).
Results
Enhancer-like regulatory elements define tissue-specific signatures
We leveraged the cell type–agnostic registry of candidate cis-regulatory elements (cCREs) generated for the human genome (hg19) by the ENCODE Project. We focused on the set of 991,173 cCREs classified as enhancer-like signatures (ELSs), defined as DNase I hypersensitive sites supported by the H3K27ac epigenetic signal, and assessed their presence–absence patterns across 43 adult cell type–specific catalogs (Supplemental Table 1; see Methods). We first explored the data with multidimensional scaling (MDS), which uncovered tissue-specific presence-absence patterns (Supplemental Fig. 1A). Indeed, the separation of samples driven by ELSs’ activity is comparable to the one obtained from the analysis of Genotype-Tissue Expression (GTEx) data (Melé et al. 2015), with blood and brain as the most diverging tissues. This suggests a correlation between gene regulatory mechanisms orchestrated by ELSs and tissue-specific gene expression patterns, which has been previously described (Pennacchio et al. 2007; Ernst et al. 2011).
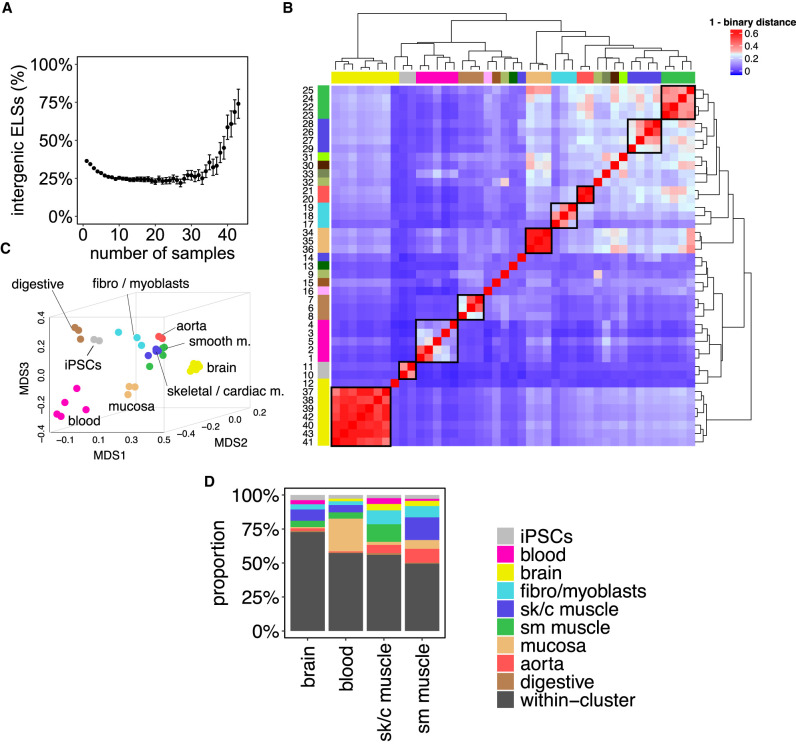
We observed that the proportion of active ELSs located in intergenic regions increases with the number of samples in which ELSs are active (Fig. 1A), suggesting an unexpected role for the genomic location of ELSs. Thus, to untangle the relationship between the genomic location and cell type–specificity of ELSs, we selected a subset of 33 samples that formed nine main tissue groups, supported by both hierarchical clustering and MDS proximity: brain, iPSCs, blood, digestive system, intestinal mucosa, fibro/myoblasts, aorta, skeletal/cardiac muscle, and smooth muscle (Fig. 1B,C; Supplemental Table 1, Samples’ Cluster). Tissues represented by only one sample (ovary, thyroid gland, lung, esophagus, spleen), or samples that do not cluster consistently with their tissue of origin and function (endocrine pancreas, liver, right lobe of liver, gastrocnemius medialis, bipolar neuron), were not included in the subsequent analyses (Supplemental Table 1; see Methods).
Figure 1.
Active enhancers define tissue identity. (A) Highly shared ELSs are more frequently located in intergenic regions. The scatterplot represents the proportion of intergenic ELSs active in increasing numbers of human adult samples. Error bars represent the 95% confidence interval. (B) Samples’ clustering defined by ELSs’ presence–absence patterns. The heat map depicts the binary distance between any pair of samples, based on the activity of 921,166 distal ELSs (±2 kb from any annotated TSS). The correspondence between samples and numbers is reported in Supplemental Table 1. (C) MDS distribution of human adult samples defined by ELSs’ activity. Analogous representation to Supplemental Figure 1A for the subset of 33 selected adult human samples. (D) Tissue-specific ELSs. The bar plot represents the type of samples found within sets of brain-, blood-, and muscle-specific ELSs. Most tissue-specific ELSs are only active in the samples of the corresponding cluster (“within-cluster”, black), but a few of them may be active in, at most, one outer sample (i.e., a sample that does not belong to the tissue cluster, colored). iPSCs-, fibro-/myoblasts-, digestive-, mucosa-, and aorta-specific ELSs are not represented, because we did not allow outer samples, given their small cluster sizes (see Methods). (sk/c) skeletal/cardiac, (sm) smooth.
The fact that tissue-specific enhancer signatures contribute to the ad hoc tissues’ functional clustering suggests a direct link between ELSs’ activity and the regulation of tissue-specific functions (Fig. 1C). Thus, we set out to characterize tissue-specific enhancer signatures and to compare them with regulatory mechanisms that are common, that is, shared among most tissues. Tissue-specific ELSs (Ts ELSs) were defined as those ELSs active in ≥80% of the samples within a given cluster and in no more than one sample outside the cluster (Supplemental Table 2; see Methods). For clusters with limited sample number (≤3), we required Ts ELSs to be active exclusively within the corresponding tissue cluster (see Methods). The overlap of Ts ELSs with samples from other clusters (Fig. 1D) is consistent with the samples’ MDS proximity observed in Figure 1C, suggesting a functional relevance of the genes regulated by shared ELSs. In addition, we identified a set of 555 ELSs active in 95% of the 33 samples, herein named as common ELSs (Supplemental Table 2).
The genomic locations of regulatory elements correlate with their tissue-homeostatic functions
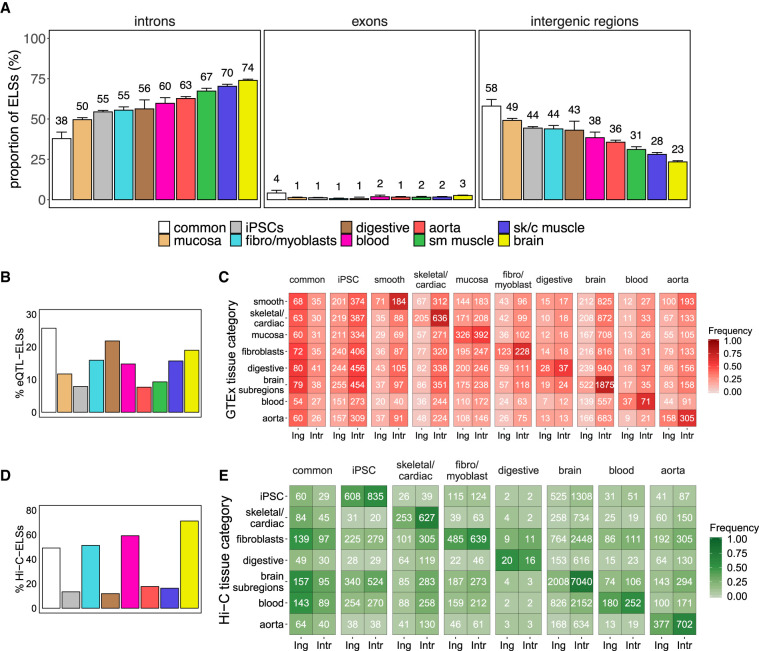
We next explored the genomic location of the sets of common and Ts ELSs. Although common ELSs are preferentially located in intergenic regions (58%) (Fig. 2A), the majority of aorta-, muscle-, and brain-specific ELSs fall inside introns (between 63% and 74%) (Fig. 2A). These significant differences in genomic distribution between tissue-specific and common regulatory elements (Supplemental Table 3) are consistent with our initial observation of a high sharing rate of intergenic ELSs across samples (Fig. 1A). In contrast, the iPSCs, fibro/myoblasts, mucosa, digestive, and blood clusters—which comprise undifferentiated, nonspecialized, highly proliferative or more heterogeneous cell types—show a more even distribution of Ts ELSs between intergenic and intronic regions (Fig. 2A). Overall, we observed a limited abundance of exonic ELSs (Fig. 2A; Supplemental Tables 3, 4).
Figure 2.
Intronic location of tissue-specific ELSs. (A) Proportions of common and tissue-specific ELSs, identified in the 33 selected human adult samples that overlap intronic, exonic, and intergenic regions. Error bars represent the 95% confidence interval. (sk/c) skeletal/cardiac, (sm) smooth). (B) Proportion of eQTL-ELSs with respect to the total amount of ELSs in each cluster. (C) Number of intergenic (Ing) and intronic (Intr) cluster-specific ELSs harboring eQTLs detected in the analyzed GTEx tissue samples. Common and iPSCs-specific ELSs were annotated with a composition of tissue-specific significant eQTLs (see Methods). Colored cells represent the proportion of region-specific eQTL-ELSs over the total amount of eQTL-ELSs per cluster. Significant differences were observed between common and tissue-specific annotated eQTL-ELSs (χ2test P-value ≤ 0.05), showing that common annotated ELSs are highly associated with intergenic regions. (D) Proportion of Hi-C-ELSs with respect to the total amount of ELSs in each cluster. (E) Number of intergenic and intronic cluster-specific ELSs overlapping Hi-C-based detected fragments in the analyzed Hi-C tissue samples. Common ELSs were annotated with a composition of tissue-specific significant Hi-C fragments (see Methods). Colored cells represent the proportion of Hi-C-ELSs over the total amount of tissue-specific Hi-C-ELSs per cluster. Significant differences were observed between common and noncommon annotated Hi-C-ELS (χ2 test P-value ≤ 0.05).
Genes harboring Ts ELSs may present distinctive features, including differences in gene and intron length. To rule out any bias in our analyses, we compared these features between genes hosting common and Ts ELSs. Although the number of introns per hosting gene is comparable across groups (Kruskal-Wallis P-value test = 0.08), we reported significant differences in gene and median intron length among tissues (Kruskal-Wallis P-value test <2.2 × 10−16) (Supplemental Fig. 1B). Nevertheless, we did not observe a correlation between such differences and the presence of intronic ELSs (Supplemental Fig. 1B). Across all tissues, most of the intronic Ts ELSs are located further than 5 kb from annotated TSSs (Supplemental Fig. 1C) and do not show chromatin marking typical of promoters (see Methods section “Tissue-specific and common ELSs”).
We subsequently explored whether the genes harboring tissue-specific intronic ELSs perform functions associated with maintenance of tissue homeostasis and response to stimuli. Indeed, the enrichment of Gene Ontology (GO) terms associated with tissue-specific cellular components is consistent with the ELSs’ tissue identity (Supplemental Table 5). For instance, genes hosting brain-specific ELSs perform functions associated with synapses and axons, whereas in the case of muscle and blood, we found significant terms related to sarcolemma, actin cytoskeleton and contractile fibers, and immunological synapses and cell membranes, respectively. Conversely, genes harboring common ELSs reported terms related to ordinary cell functions and membrane composition (Supplemental Table 5). Although this suggests an implication of intronic ELSs in tissue-specific functions, likely through tissue-specific gene regulation mechanisms, there is no proven evidence of intronic ELSs being direct regulators of their host genes. To identify genes targeted by Ts ELSs, we integrated our ELS analysis with the catalog of expression quantitative trait loci (eQTLs) provided by the Genotype-Tissue Expression Project (The GTEx Consortium 2017). eQTLs provide functional information about the changes of expression associated with human variants. We leveraged eQTLs located in both intronic and intergenic ELSs to identify their target genes. Among the 48,555 common and Ts ELSs, 6349 overlap with a significantly associated eQTL-eGene pair, hereafter referred to as eQTL-ELSs. The proportion of eQTL-ELSs is similar among the tissue samples represented in the GTEx sampling collection, ranging between 10% and 25% (Fig. 2B). In all annotated tissues, gene regulation driven by eQTL-ELSs occurs predominantly in the tissue where the ELS is specifically active (Fig. 2C). In line with the above-mentioned results (Fig. 2A), highly specialized tissues such as brain and muscle show the highest proportion of intronic versus intergenic ELSs hosting eQTLs detected in the corresponding tissue (Fig. 2B,C). Conversely, common eQTL-ELSs are more frequently located in intergenic elements (32% vs. 62%) (Fig. 2C). GO enrichment analysis on the sets of target genes associated with intronic and intergenic eQTL-ELSs shows a clear prevalence of tissue-specific terms for those genes targeted by intronic rather than intergenic eQTL-ELSs—for instance, skeletal/cardiac muscle: carbohydrate and amino acid metabolism; brain: cell projection and microtubule cytoskeleton organization (Supplemental Table 6). In contrast, genes associated with common eQTL-ELSs (either intronic or intergenic) do not show any significantly enriched term. Altogether, these results suggest that intronic eQTL-ELSs are involved in the regulation of genes associated with tissue-specific functions, whereas intergenic ELSs are more devoted to tissue homeostatic processes.
Target genes of intronic ELSs identified by Hi-C regulate tissue-specific functions
The interaction between ELSs and promoters is central for the onset of gene expression. These types of interactions are defined in each tissue and can be identified genome-wide through Hi-C-seq. Here, we explored ELS-promoter interactions reported by published Hi-C data sets in relevant tissues, identifying Ts ELS-target genes, and thus improving the annotations of ELSs-target genes with respect to the eQTL analysis (Fig. 2D,E; Mifsud et al. 2015; Jung et al. 2019; Lu et al. 2020). This approach allowed us to observe that most of the target genes are regulated by multiple ELSs (Supplemental Fig. 2A). As in the case of eQTL-ELSs, brain and muscle show the highest proportion of intronic versus intergenic ELSs intersecting Hi-C interacting fragments in the corresponding tissue, whereas common Hi-C-ELSs are enriched in intergenic regions (Fig. 2E). The GO enrichment analysis reported an increase in relevant terms involved in tissue-specific functional roles as well. Of note, intronic Hi-C-ELSs show stronger enrichment in tissue-specific terms (skeletal/cardiac muscle: I band and Z disc components; brain: pre-/postsynaptic assembly and organization; aorta: regulation of smooth muscle cell migration and proliferation), whereas we observed broader functionality from intergenic ELSs’ interactions (brain: choline catabolic process and copper ion homeostasis, among others) (Supplemental Table 7). Moreover, common Hi-C-ELSs appear to target genes that are enriched in housekeeping functions, such as cell adhesion and nucleosome organization (Supplemental Table 7). Overall, these results on ELS-promoter interactions further support the fact that intronic ELSs regulate genes controlling tissue-specific functions, whereas intergenic ELSs are more devoted to tissue homeostatic processes.
Intronic ELSs regulate the expression of hosting and non-hosting genes
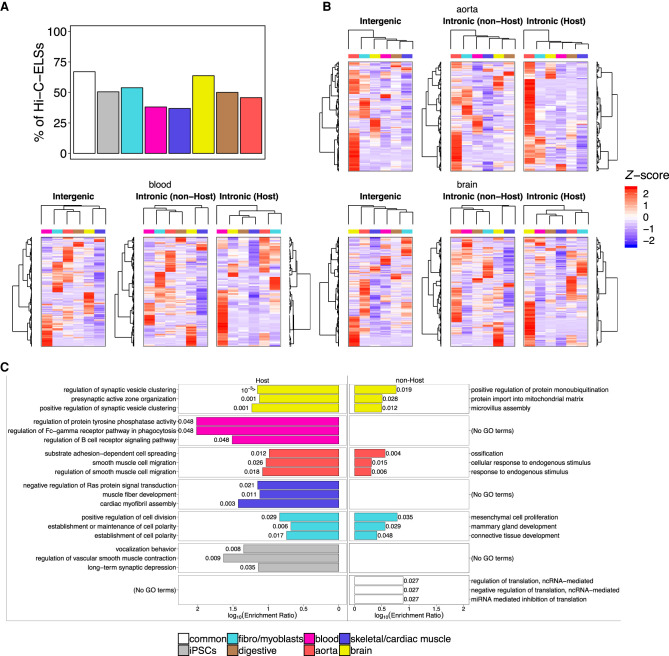
Next, we wanted to understand the relationship between tissue-specific intronic ELSs and their host genes. To do so, we analyzed the expression patterns of genes targeted by Hi-C ELSs, as a proxy for direct regulation. The proportion of intronic Hi-C-ELSs targeting their host genes is comparable among most groups of samples and ranges between 45% and 65%, with the exception of muscle and blood, which show lower values (Fig. 3A). We compared the expression patterns of Hi-C-ELSs’ target genes, considering the type of ELS regulating them (intergenic; intronic host—i.e., an ELS targeting its host gene; intronic non-host—i.e., an ELS targeting a gene that is not its host gene). Genes regulated by intronic host ELSs exhibit expression patterns that better recapitulate tissue identity (the relevant tissue clusters, in almost all the cases, separately from the other groups), whereas hierarchical clustering of genes regulated by intronic non-host ELSs does not efficiently discriminate tissue-specific patterns (Fig. 3B; Supplemental Fig. 2B). Genes regulated by intronic host ELSs are associated with tissue-specific functions (Supplemental Table 8), in particular synaptic vesicle clustering and active zone organization for brain (e.g., PCDH17), regulation of cell division and establishment of cell polarity for fibroblasts (e.g., TGFB2), cardiac myofibril assembly and muscle fiber development for skeletal/cardiac muscle (e.g., MEF2A), and regulation of smooth muscle cell migration for aorta (e.g., DOCK5). On the contrary, those genes targeted by intronic non-host ELSs are involved in homeostatic functions not uniquely associated with the relevant tissue, suggesting that they are not expressed in a tissue-specific manner but are nevertheless regulated by tissue-specific enhancers. For instance, brain and aorta present significant terms related to protein monoubiquitination (e.g., PDCD6) and cellular response to endogenous stimulus (e.g., TNC), respectively (Fig. 3C). Overall, this indicates that the intronic location of regulatory elements cannot be associated exclusively with the regulation of the host gene. Furthermore, the identification of a large proportion of intronic non-host ELSs suggests that the intronic location may be, in a particular tissue, advantageous for the establishment and maintenance of gene expression programs, including non-tissue-specific events.
Figure 3.
Intronic enhancers regulate hosting and non-hosting genes. (A) Proportions of Hi-C-ELSs that target their host gene. These proportions were calculated over the total amount of intronic Hi-C-ELSs within each cluster. (B) Z-score normalized median gene expression levels, across GTEx tissue categories, of the genes targeted by intergenic and intronic Hi-C-ELSs. Intronic Hi-C-ELSs are distinguished between those targeting their host gene (Host) and those that target a gene outside their hosting region (non-Host). Dendrograms show the hierarchical clustering of target genes (rows) and GTEx tissue categories (columns). (C) Top three significantly enriched GO terms found in the genes targeted by host and non-host intronic Hi-C-ELSs. P-values (FDR corrected) are shown for each enriched term.
The enrichment of transcription factor binding sites in Ts ELSs is independent of their genomic location
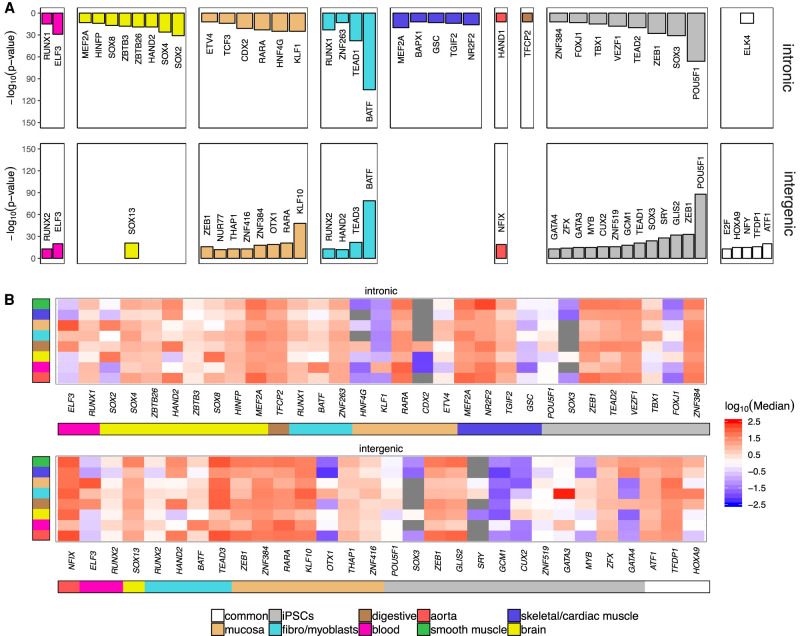
The activation of ELSs is a dynamic process depending on, among other factors, its accessible chromatin to be bound by transcription factors (TFs). Thus, tissue-specific gene expression programs may be controlled by the underlying signature of TFs-ELSs pairing (Schmitt et al. 2016). We next wondered whether the specific distribution of ELSs, that is, intronic versus intergenic, is associated with a different transcription factor binding site (TFBS) signature that could account for their tissue-specific activity. To this purpose, we explored, with HOMER (Heinz et al. 2010), the enrichment of TFBSs independently for intronic and intergenic ELSs. Indeed, a distinct TFBS signature for each tissue in both intronic and intergenic ELSs can be observed (Fig. 4A), supporting our previous results that Ts ELSs significantly contribute to the regulation of tissue-specific functions. The number of enriched TFBSs in intronic regions is higher in highly specialized tissues such as brain and muscle and shows no overlap with TFBSs found in intergenic ELSs. The opposite picture is observed in common ELSs, with higher enrichment of TFBSs in intergenic ELSs. An intermediate pattern is observed for highly proliferative tissues such as iPSCs, fibroblasts, mucosa, and blood, in which the amount of enriched TFBSs is similar between intronic and intergenic ELSs (Fig. 4A; Supplemental Table 9). Among the TFBSs enriched in Ts intronic and intergenic ELSs, we found well-known TFs associated with tissue-specific homeostatic events, such as RUNX2 in blood controlling adult endothelial hemogenesis (Lis et al. 2017) and SOX4 and SOX8 in brain controlling adult neural differentiation (Chen et al. 2015). POU5F1 (previously known as OCT4) is required for iPSCs. Still, with the exception of those TFs associated with enriched TFBSs in iPSCs, most other TFs are widely expressed across tissues (Fig. 4B). This distinct iPSCs’ TF-ELS binding potential is supported by previous data indicating that iPSCs share their epigenetic signature with early developmental stages rather than with the original tissue prior to reprogramming. Overall, the TFBSs’ enrichment, rather than the TFs’ gene expression patterns, is the most variable feature between intronic and intergenic ELSs and among tissues.
Figure 4.
Differential TFs programs activate intronic and intergenic ELSs in a tissue-specific manner. (A) Bar plots reporting the significantly enriched TFBSs in intronic and intergenic tissue-specific ELSs. (B) Z-score normalized median gene expression, across GTEx tissue categories, of the TFs that bind to significantly enriched TFBSs in each group.
Dynamic location of ELSs throughout embryonic development and maturation
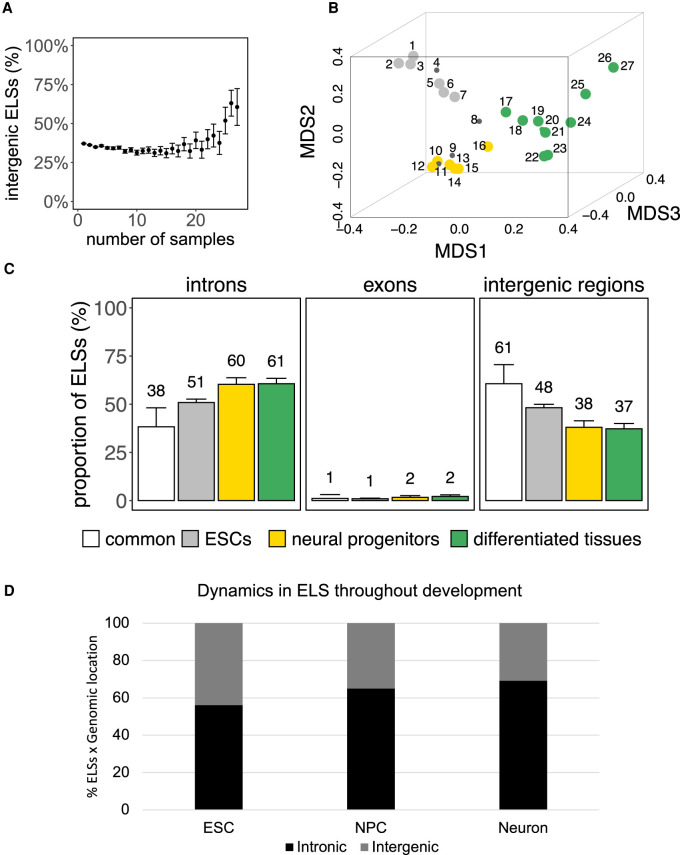
Throughout embryonic development, tissues mature to fully reach their functional capacity in adulthood, giving rise to several tissue-specific homeostatic features that vary among different tissues. For instance, blood comprises a wide number of cell types characterized by heterogeneous functions and high turnover. On the opposite side, we find highly specialized tissues such as muscle, that are formed by fewer cell types, mainly dedicated to the same function and with limited cell division capacity. During development, tissues share features of basic homeostasis, proliferation, and plasticity, but they are also already patterned to perform their adult functions. Still, whether the regulatory features of a given adult tissue are reminiscent of their developmental lineage remains largely unknown. To answer this question, we assessed the activity and the intronic location of the 991,173 cell type–agnostic ELSs across 27 embryonic samples (Fig. 5A; Supplemental Table 10). MDS analysis highlighted three main groups of embryonic samples: stem cells (ESCs), neural progenitors, and a larger group of more differentiated cell types (Fig. 5B; Supplemental Table 10, Samples’ Group). The three groups of samples are associated with 3112, 784, and 1166 specific ELSs, respectively (Supplemental Table 11). Although the majority of these ELSs are active only within the corresponding cluster, we reported that 26% of the neural progenitors-specific ELSs are also active in one ESCs sample (Supplemental Fig. 3A). On the contrary, we identified only 94 ELSs common to all embryonic samples (Supplemental Table 11). The proportion of specific intronic ELSs is higher for neural progenitors and differentiated tissues, compared to ESC-specific and common ELSs (Fig. 5C), but lower with respect to clusters of adult tissues such as aorta, muscle, and brain (Fig. 2A). As in the case of adult samples, we observed a limited abundance of exonic ELSs (Fig. 5C; Supplemental Tables 12, 13), whereas we could not find significant associations between the frequency of group-specific intronic ELSs and features of gene and intron length (Supplemental Fig. 3B). As for adult samples, most of these group-specific intronic ELSs are located further than 5 kb from annotated TSSs (Supplemental Fig. 3C).
Figure 5.
Dynamic localization of ELSs throughout embryonic development. (A) Correlation between ELSs’ sharedness among embryonic samples and frequency of their intergenic localization (error bars represent the 95% confidence interval). (B) MDS representation of embryonic samples defines three main groups of tissues (ESCs in gray; neural progenitors in yellow; more differentiated tissues in green). (C) ELSs specific to neural progenitors and differentiated tissues are more frequently intronic, whereas common ELSs are preferentially intergenic. (D) Dynamics of the localization of active ELSs during ESC-derived maturation stages (hESC, neural progenitors, and neurons). ELSs defined by H3K27ac+/H3K4me3− peaks during ESC-derived neural maturation, that also overlap with ENCODE ELSs, increasingly distribute in intronic regions as maturation advances.
Next, we wanted to validate the dynamics of intronic versus intergenic ELSs active throughout development, using brain development as a paradigm (Supplemental Fig. 4A). To this purpose, we identified active ELSs (ChIP-seq H3K27ac + /H3K4me3- peaks) in human ESCs, and hESC-derived NPCs and neurons, and assessed their degree of overlap with ENCODE ELSs. Active ELSs identified by ChIP-seq in ESCs, NPCs, and neurons overlap with ENCODE ELSs specific to ESCs (86%), embryonic neural progenitors (40%), and adult brain (53%) samples, respectively. In particular, the proportion of active intronic ELSs increases with the degree of differentiation of the samples (55% in ESCs, 64% in NPCs, and 68% in neurons) (Fig. 5D), validating the observed correlation between active Ts ELSs and their intronic location. We observed a high overlap (86% to 98%) between ENCODE common embryonic ELSs and H3K27ac ChIP-seq peaks detected during the hESC-differentiation, including known ELSs for housekeeping genes, such as ACTB (Supplemental Fig. 4B). The expression of genes regulated by individual candidate ELSs (Supplemental Fig. 4C–E) is, in most cases, consistent with the activity of the ELS, being active either in a tissue-specific manner in ESCs or neurons, or in all three differentiation stages (Supplemental Table 14). Although a small fraction of common ELSs is marked by H3K4me3 in ESCs, NPCs, and neurons (Supplemental Fig. 4B), the corresponding H3K4me3 signal is comparatively lower than the H3K4me3 level observed at promoter regions (Supplemental Fig. 4F). When analyzing the genes hosting developmental group-specific intronic ELSs, we observed that they are enriched in functions consistent with the corresponding adult tissue (Supplemental Table 15). For instance, the ones hosting neural progenitors-specific ELSs are enriched in neural development-related terms, such as axonogenesis and dendritic spine organization. On the contrary, genes hosting developmental common ELSs are enriched in protein complexes like nBAF and SWI/SNF, known developmental chromatin remodelers (Alver et al. 2017).
Lastly, in an attempt to define the amount of regulatory activity shared by embryonic and adult samples as an indicator of the reminiscent embryonic function in adult tissue homeostasis, we computed, for specific and common embryonic ELSs, the number of adult tissues in which they are found active. As expected, whereas ELSs specific to ESCs and neural progenitors are active in a limited set of adult samples, embryonic differentiated tissues report a higher degree of shared regulatory activity with adult cell types (Supplemental Fig. 5). Moreover, ELSs active in all embryonic samples (common) are also active in the majority of adult samples. Overall, these results show that the genomic location of ELSs is dynamic throughout development and shifts toward an intronic localization during tissue maturation.
Discussion
In this study, we show the central role of intronic enhancer-like signatures in the control of tissue-specific expression programs. Since Heitz described in 1928 (Heitz 1928) euchromatin as transcription-permissive chromosomal regions enriched in genes, and heterochromatin as inactive or passive chromatin regions, this dual definition has been shaped throughout the years, but it still remains vastly correct (Ernst and Kellis 2010; De Laat and Duboule 2013; DeMare et al. 2013). Intergenic regions are often regulatorily silenced, and this happens more frequently in adult than embryonic tissues (Heinz et al. 2015). The ENCODE Project reports that about half of the ELSs are intergenic, and 38% are intronic (ENCODE SCREEN Portal: https://screen-v10.wenglab.org/, section “About”). In our study, we describe an enrichment of intronic ELSs in the most specialized tissues. These elements regulate genes involved in tissue-specific functions, suggesting an important role for the genomic location of ELSs. On the contrary, in less specialized adult tissues and embryonic samples, ELSs are less frequently found in intronic elements, suggesting that the maturation and tissue commitment correlates with the ELSs’ distribution across the whole genome. One could hypothesize that the enriched presence of intronic ELSs is advantageous for the control of the gene expression signature of a particular tissue, for instance, granting ELSs accessibility in open DNA regions (genes) and avoiding their leaky activity. In line with this, active transcription and nascent RNA have been recently associated with the maintenance of open chromatin (Hilbert et al. 2021), a process that can be advantageous to the presence of intronic ELSs in actively transcribed genes. Introns have long been observed as gene expression regulators through different mechanisms (Chorev and Carmel 2012; Shaul 2017; Rose 2019). Specifically, introns’ regulatory potential has been associated with the regulation of the host gene's expression in several different ways, often related to alternative splicing, intron retention (Jacob and Smith 2017), non-sense mediated decay (Lewis et al. 2003), and to the control of transcription initiation via recruitment of RNA Polymerase II, likely as alternative promoters (Bieberstein et al. 2012; Kowalczyk et al. 2012). However, here we found that, in most tissues, about half of the ELSs located in introns do not regulate the expression of the host gene but of genes involved in important tissue homeostatic functions, whose expression is not restricted to that particular tissue. This is important regulatory information, because it disentangles the presence of intronic ELSs from the regulation of the host gene, opening new opportunities to identify the regulatory mechanisms controlling tissue-specific gene expression. Overall, our results suggest that the genomic distribution of tissue-specific active ELSs is not stochastic and mainly overlaps with intronic elements. The opposite happens to active ELSs common to all tissues, which are instead enriched in intergenic regions. These results suggest that intronic enhancers play a role in the regulation of gene expression in a tissue-specific manner.
Methods
The ENCODE registry of candidate cis-regulatory elements
The cell type–agnostic registry of human candidate cis-regulatory elements available from the ENCODE portal corresponds to a subset of 1,310,152 representative DNase I hypersensitivity sites (rDHSs) in the human genome with epigenetic activity further supported by histone modification (H3K4me3 and H3K27ac) or CTCF-binding data (https://screen-v10.wenglab.org/; section “About”). It comprises 991,173 enhancer-like signatures, 254,880 promoter-like signatures (PLSs), and 64,099 CTCF-only signatures. In addition, cell type–specific catalogs are provided for those cell types with available DNase and ChIP-seq ENCODE data.
Selection of cCREs with enhancer-like signature across human samples
We downloaded the set of 1,310,152 cell type–agnostic cCREs for human assembly 19 (hg19) from the ENCODE SCREEN webpage (https://screen-v10.wenglab.org/; file ID: ENCFF788SJC). From the ENCODE portal (www.encodeproject.org/matrix/?type=Annotation&encyclopedia_version=ENCODE+v4&annotation_type=candidate+Cis-Regulatory+Elements&assembly=hg19), we retrieved cell type–specific registries of cCREs for 43 adult and 27 embryonic human samples with available DNase data and ChIP-seq H3K4me3 and H3K27ac data. The ENCODE File Identifiers for the adult and embryonic data sets are reported in Supplemental Tables 1 and 8, respectively. No significant changes are expected upon realignment to GRCh38, because main improvements with respect to hg19 have been made in the representation of so-called alternate haplotypes, with a small impact on the definition of genic and intergenic regions (Church et al. 2015). We focused on the 991,173 cell type–agnostic cCREs with ELS activity and generated a binary table in which we assessed, for a given cCRE, the presence/absence of ELS activity annotation (column 9 = “255, 205, 0”) in each of the 43 adult and 27 embryonic samples. A binary distance matrix between all pairs of adult samples was used to perform multidimensional scaling in three dimensions. This resulted in the selection of 33 adult samples, which form nine tissue groups well supported by hierarchical clustering (Fig. 1B,C) The same procedure was applied, independently, to the embryonic samples. In this case, IMR-90, mesendoderm, mesodermal cell, endodermal cell, and ectodermal cell samples were not included in subsequent analyses.
Intersection of ELSs with genes, introns, exons, and intergenic regions
Genes’, exons’, and introns’ coordinates were obtained from GENCODE v19 annotation (https://www.gencodegenes.org/human/release_19.html). The overlap between ELSs and genes, exons, and introns was computed using BEDTools intersectBed v2.27.1 (Quinlan and Hall 2010). The proportions of ELSs overlapping intronic segments (Figs. 2A, 5C) also include a limited set of ELSs overlapping both intronic and exonic regions. On the other hand, we defined as exonic ELSs those intersecting exclusively exonic regions (Figs. 2A, 5C). The overlap of ELSs with intergenic regions was obtained by intersecting the former with the genes’ coordinates using the BEDTools intersectBed option -v.
Tissue-specific and common ELSs
Tissue-specific ELSs are ELSs active (see Methods section “Selection of cCREs with enhancer-like signature across human samples”) in ≥80% of the samples within a given group of samples (blood = 4/5; skeletal/cardiac muscle = 3/4; smooth muscle = 3/4; brain = 6/7; stem cells = 5/6; neural progenitors = 5/6; differentiated tissues = 8/10). Because of the small sample size, we required iPSCs-, fibro-/myoblasts-, digestive-, mucosa-, and aorta-specific ELSs to be active in 100% of the samples (either 2/2 or 3/3). In addition, Ts ELSs are active in 0 (iPSCs, fibro-/myoblasts, digestive, mucosa, and aorta) or at most one (all other groups) outer sample (i.e., samples outside of the considered group). Common adult and embryonic ELSs are ELSs active in 95% and 100% of the samples, respectively (i.e., 31/33 and 22/22). To rule out indirect effects of ELS activity related to promoter regions, we discarded common and Ts ELSs overlapping any annotated transcription start site (TSS, ±2 kb) in GENCODE v19. We further computed, for every tissue-specific intronic ELS, the minimum distance from any annotated TSS (Supplemental Figs. 1C, 3C): most of these ELSs are located more than 5 kb from TSSs. We also controlled our sets of Ts ELSs for the presence of potential alternative promoters, leveraging every adult and embryonic sample with available H3K4me3 ChIP-seq experiments from ENCODE (Supplemental Fig. 6; Supplemental Tables 16, 17). More specifically, we computed the proportion of intronic and intergenic Ts ELSs showing peaks of H3K4me3 (Supplemental Fig. 6A,B). Overall, we did not observe differences in the proportion of marked regions between intronic and intergenic ELSs. In the case of marked ELSs, we compared, for both adult and embryo samples, their aggregated H3K4me3 signal (expanding ±5 kb from the center of the ELS) to the signal detected at marked annotated TSSs (±2 kb) (for some examples, see Supplemental Figs. 6C,D, 7).
Assessing enhancer regulatory activity with GTEx eQTL-eGene significant pairs
ELSs were annotated using the GTEx v7 (The GTEx Consortium 2017) significant variant-gene pairs from 46 different tissues (number of samples with genotype ≥70), available on the GTEx portal (www.gtexportal.org). Only single-tissue eQTL-eGene associations with a qval ≤ 0.05 were used. Similar GTEx tissues were grouped in unique categories in order to consider the most complete catalog of eQTL-eGene pairs per group of samples. These categories were named as follows: fibroblasts (Skin Not Sun Exposed Suprapubic, Cells Transformed Fibroblasts), blood (Whole Blood, Spleen), skeletal/cardiac muscle (Skeletal Muscle, Heart Atrial Appendage, Heart Left Ventricle), brain subregions (all brain subregions, Pituitary Gland, Nerve Tibial), Aorta (Artery Aorta), smooth muscle (Artery Coronary, Artery Tibial), digestive (Liver, Pancreas, Small Intestine Terminal Ileum, Stomach, Colon Sigmoid, Colon Transverse, Esophagus Gastroesophageal Junction, Esophagus Muscularis, Adipose Subcutaneous, Adipose Visceral Omentum), mucosa (Esophagus Mucosa), gland (Adrenal Gland, Thyroid, Minor Salivary Gland), breast (Breast Mammary Tissue), lung (Lung), sexual tissues (Ovary, Prostate, Testis, Uterus, Vagina). BEDTools (Quinlan and Hall 2010) was used to intersect the Ts ELSs’ coordinates with the cis-eQTLs’ positions in the considered genomic locations (intronic and intergenic). We kept all eQTL-eGene pairs that were found significantly associated with the matching eQTL-ELS's tissue category (muscle skeletal/cardiac, muscle smooth, fibro-/myoblast, digestive, mucosa, brain, blood, aorta). In the case of iPSCs-specific and common ELSs, we considered those eQTL-eGene pairs that were significantly reported in at least 50% of all the tissues. The resulting intersected ELSs were considered as being responsible for the regulation of the associated eGene. The functional enrichment of the ELSs’ target genes was performed by the online utility WebGestalt (Liao et al. 2019).
Assessing enhancer regulatory activity with Hi-C-based significant fragment pairs from loop contacts
ELSs were also annotated using significant Hi-C-based interacting fragment pairs from three independent data sets (Mifsud et al. 2015; Jung et al. 2019; Lu et al. 2020). Different primary tissue and cell line samples were used to annotate each of the Ts ELSs categories in our study, except for smooth muscle, for which no Hi-C samples were found. As for the GTEx samples’ groups in the previous section, we grouped the Hi-C samples in unique categories in order to consider the most complete catalog of Hi-C fragment pairs per group of samples. These categories were named as follows: skeletal/cardiac muscle (Right ventricle [RV], Right heart atrium [RA3], Psoas [PO3], left ventricle [LV]), fibro-/myoblasts (Fibroblast cells [IMR-90]), brain (Hippocampus, dorsolateral prefrontal cortex, cortex adult, Neuron), blood (GM12878 + GM19240 lymphoblastoid cell line, CD34, GM12878), iPSCs (iPSCs), aorta (Aorta), mucosa (Sigmoid Colon), digestive (Pancreas, Gastric tissue). In order to identify the significant ELS-gene pairs, BEDTools (Quinlan and Hall 2010) was used to intersect the Hi-C fragment coordinates with our ELSs associated with the different genomic locations (intronic and intergenic). In those cases in which the other fragment did not belong to any other ELS, we intersected them with the GENCODE annotation (v19), inferring in this way the target genes of these ELSs. As for the eQTL annotation, only the Hi-C–based ELS-gene interactions associated with the matching Hi-C–ELSs’ tissue category were kept (iPSC, skeletal/cardiac muscle, fibro-/myoblast, digestive, brain, blood, aorta). Mucosa- and smooth muscle-Ts ELSs were removed from the analysis due to the lack of intersection with significant fragment pairs and Hi-C sample tissues, respectively. In the case of common ELSs, we considered the ELS-gene pairs reported in at least 50% of all the Hi-C tissue samples. After the annotation of our ELSs, we ended up with a collection of enhancer-gene interactions where the target gene was considered as being regulated by the interacting ELS. In order to define the sets of intronic Host/non-Host ELSs in Figure 3A, we identified the ELSs’ target genes that are also the host gene of that ELS. If a particular ELS presents among their target genes also its own host gene, then that ELS was classified as Host, if none of the target genes is hosting the ELS, then that element was classified as non-Host. When considering the interactions ELS-gene in Figure 3B and Supplemental Figure 2B, we defined an interaction as Host if the target gene is hosting that ELS; otherwise, if the same ELS is targeting a gene that is not hosting the element, that interaction is classified as non-Host. The target gene expression values were obtained from the GTEx expression data (v7), and Z-score normalized across the different GTEx tissue categories. The hierarchical clustering analyses of the Host/non-Host target genes and GTEx tissue categories were performed with the R function hclust. The functional enrichment analyses on the ELSs’ target genes and Host/non-Host target genes were performed with the online utility WebGestalt (Liao et al. 2019).
cis-regulatory elements and transcription factor binding sites
Transcription factor binding sites were predicted by using the motif discovery software HOMER (Heinz et al. 2010). This program performs a differential motif discovery by taking two sets of genomic regions (findMotifGenome.pl script) and identifying the motifs that are enriched in one set of sequences relative to a background list of regions. We analyzed the Ts ELSs’ binding motifs by considering the ELS regions from all the other tissues as background. We searched for 6-mer- and 7-mer-length motifs as a way to focus on enriched core motif sequences and avoid redundancy from longer motifs with similar functions. A hypergeometric test and FDR correction were applied for the motif enrichment. Only significantly enriched motifs were considered in the subsequent analyses. The functionality of the predicted TFBSs was assessed by analyzing the tissue-specific expression of the transcription factors that bind to them. GTEx expression data (v7) were analyzed for those transcription factors whose TFBSs were reported as significant by HOMER in all tissues and genomic locations. In the gene expression analysis, some transcription factors were removed due to the lack of expression data. Z-score normalization was performed across the different GTEx tissue categories in all transcription factors.
ChIP-seq data generation and processing
ChIP-seq was performed in hESC line H9 (WiCell), hESC-derived neural progenitors (NPCs), and neurons. hESCs were maintained in culture in mTESR (Stem Cell Technologies), and NPCs and neurons were obtained upon cerebral organoid differentiation (Lancaster and Knoblich 2014). Briefly, 9000 H9 hESCs were seeded in a low attachment 96-well plate (Corning) with ROCK inhibitor in mTESR. After 6 d, neuroepithelium differentiation was triggered using induction media for another 6–8 d, until the neuroepithelium was detectable, and subsequently transferred to the neural expansion in Matrigel (Corning). Organoids were disaggregated at day 30 postdifferentiation and maintained in neural differentiation media (common N2B27) supplemented with 20 ng/mL of each EGF (Thermo Fisher Scientific PHG0315) and FGF2 (Peprotech 100-18B) to obtain a 2D NPC monolayer. NPCs were harvested after two passages. Neurons were terminally differentiated in maturation media (N2B27) for three more weeks. Cells were harvested with Cell Dissociation Solution (Stem Cell Technologies) and kept at −80°C. DNA was crosslinked with formaldehyde for 10 min at room temperature. Fixation was stopped by incubating with PBS/0.1% Triton X-100/0.125 M glycine for 5 min at room temperature, and chromatin was fragmented in a Q-sonica sonicator (15 min constant sonication at 40% amplitude). H3K27ac (Active Motif reference 39336) and H3K4me3 (Active Motif 39916) antibodies were used for immunoprecipitation following the protocol previously described (Pérez-Lluch et al. 2015). ChIP libraries were performed following Illumina procedures. Libraries were quantified by Qubit (Thermo Fisher Scientific) and visualized in a fragment analyzer (Agilent) previous to sequencing. Sequencing was performed in an Illumina NextSeq 500, single-end run, following the instructions of the manufacturer.
Data were processed using the ChIP-nf (https://github.com/guigolab/chip-nf) Nextflow (Di Tommaso et al. 2017) pipeline. Input samples were downsampled to a number of reads comparable to the ChIP samples with the tool seqtk (https://github.com/lh3/seqtk). ChIP-seq reads were aligned to the human genome assembly (GRCh37) using the GEM (Marco-Sola et al. 2012) mapping software, allowing up to two mismatches. Only alignments for reads mapping to 10 or fewer loci were reported. Duplicated reads were removed using Picard (http://broadinstitute.github.io/picard/). Peak calling was performed using Zerone (Cuscó and Filion 2016) with replicates handled internally. Pile-up signal from bigWig files was obtained running MACS2 (Zhang et al. 2008) on individual replicates. No shifting model was built. Instead, fragment length was defined for each experiment and used to extend each read toward the 3′ end (using the ‐‐extsize option). Pile-up signal was normalized by scaling larger samples to smaller samples (using the default for the ‐‐scale-to option) and adjusting signal per million reads (enabling the ‐‐SPMR option). To calculate the proportion of ELSs described in Figure 5D, only active candidate ELSs (H3K27ac+/H3K4me3−) overlapping ENCODE tissue-specific ELSs in the matched ENCODE biosamples were considered (i.e., neuron ELSs overlapping ENCODE adult brain-specific ELSs; NPCs ELSs overlapping ENCODE neural progenitors-specific ELSs; ESCs ELSs overlapping ENCODE ESCs-specific ELSs).
Gene expression analysis
To validate gene expression regulation, target genes regulated by intronic or intergenic ELSs were selected based on the following criteria (see also Supplemental Table 14): (1) controlled by a single ENCODE ELS in adult samples, either brain-specific or common (column “Tissue”); (2) showing H3K27ac+/H3K4me3− peaks in the relevant cell's ChIP-seq validation (column “Peak ChIP-seq”); and (3) not overlapping with exons.
RNA was obtained from hESCs, NPCs, and neuron pellets used for ChIP-seq. Retrotranscription was performed using SuperScript III retrotranscriptase. qPCR was performed in 10 ng cDNA with the Roche SYBR Green Master Mix. Primers used for qPCR are reported in Supplemental Table 14. Gene expression is reported following the relative expression of the DDCt method. GAPDH and ACTB were used as reference genes. ACTB gene expression showed more stability throughout the differentiation process and, therefore, it was used as the reference gene for the analysis.
Statistical analyses and visualization
All statistical analyses and visualization plots were performed using the R language for statistical computation and graphics (https://www.R-project.org/) (R Core Team 2017).
Data access
All raw and processed sequencing data generated from this study have been submitted to ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-10595.
Supplementary Material
Acknowledgments
S.A. is a Serra-Hunter Fellow since 2021. S.A. is supported by a fellowship from the Secretaria d'Universitats i Recerca del Departament d'Empresa i Coneixement (Generalitat de Catalunya) (BP-2017-00176). B.B. is supported by the fellowship 2017FI_B00722 from the Secretaria d'Universitats i Recerca del Departament d'Empresa i Coneixement (Generalitat de Catalunya) and the European Social Fund (ESF). P.V-M. is supported by an FPI PhD fellowship (FPI-BES-2016-077706) part of the “Unidad de Excelencia María de Maeztu” funded by the Ministerio de Economía, Industria y Competitividad, Gobierno de España (MINECO) (ref: MDM-2014-0370). J.B. is funded by PID2019-110933GB-I00/AEI/10.13039/501100011033 awarded by the Agencia Estatal de Investigación (AEI) and with the support of Secretaria d'Universitats i Recerca de la Generalitat de Catalunya (GRC 2017 SGR 702). The DCEXS at UPF is part of the “Unidad de Excelencia María de Maeztu” funded by the AEI (CEX2018-000792-M). We thank the ENCODE and GTEx Consortia for data production. We thank Diego Garrido-Martín (R. Guigó Lab) for valuable statistical advice.
Author contributions: S.A., B.B, and P.V-M. designed the study, analyzed the data, and wrote the manuscript with feedback from all the authors. S.A. and S.P-L. performed ChIP-seq experiments, I.T. generated the differentiations for the ChIP-seq. H.L. and A.S-C. performed some of the bioinformatic analyses. J.B. and R.G. contributed to editing of the manuscript. S.A. supervised the study.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.270371.120.
Competing interest statement
The authors declare no competing interests.
References
- Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, Haswell JR, Park PJ, Roberts CW. 2017. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun 8: 14648. 10.1038/ncomms14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberstein NI, Oesterreich FC, Straube K, Neugebauer KM. 2012. First exon length controls active chromatin signatures and transcription. Cell Rep 2: 62–68. 10.1016/j.celrep.2012.05.019 [DOI] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Lee GA, Pourmorady A, Sock E, Donoghue MJ. 2015. Orchestration of neuronal differentiation and progenitor pool expansion in the developing cortex by SoxC genes. J Neurosci 35: 10629–10642. 10.1523/JNEUROSCI.1663-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Yu W, Tober J, Gao P, He B, Lee K, Trieu T, Blobel GA, Speck NA, Tan K. 2019. Spatial genome re-organization between fetal and adult hematopoietic stem cells. Cell Rep 29: 4200–4211.e7. 10.1016/j.celrep.2019.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorev M, Carmel L. 2012. The function of introns. Front Genet 3: 55. 10.3389/fgene.2012.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choukrallah MA, Song S, Rolink AG, Burger L, Matthias P. 2015. Enhancer repertoires are reshaped independently of early priming and heterochromatin dynamics during B cell differentiation. Nat Commun 6: 8324. 10.1038/ncomms9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Schneider VA, Steinberg KM, Schatz MC, Quinlan AR, Chin CS, Kitts PA, Aken B, Marth GT, Hoffman MM, et al. 2015. Extending reference assembly models. Genome Biol 16: 13. 10.1186/s13059-015-0587-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuscó P, Filion GJ. 2016. Zerone: a ChIP-seq discretizer for multiple replicates with built-in quality control. Bioinformatics 32: 2896–2902. 10.1093/bioinformatics/btw336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laat W, Duboule D. 2013. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502: 499–506. 10.1038/nature12753 [DOI] [PubMed] [Google Scholar]
- DeMare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP. 2013. The genomic landscape of cohesin-associated chromatin interactions. Genome Res 23: 1224–1234. 10.1101/gr.156570.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. 2017. Nextflow enables reproducible computational workflows. Nat Biotechnol 35: 316–319. 10.1038/nbt.3820 [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2013. Human housekeeping genes, revisited. Trends Genet 29: 569–574. 10.1016/j.tig.2013.05.010 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583: 699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. 10.1038/nbt.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Sutherland H, de Las Heras J, Allan J, Jenuwein T, Bickmore WA. 2003. Formation of facultative heterochromatin in the absence of HP1. EMBO J 22: 5540–5550. 10.1093/emboj/cdg520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S. 1983. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33: 717–728. 10.1016/0092-8674(83)90014-4 [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491. 10.1016/j.stem.2010.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16: 144–154. 10.1038/nrm3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. 1928. Das heterochromatin der moose. Jahrb Wiss Botanik 69: 762–818. [Google Scholar]
- Hilbert L, Sato Y, Kuznetsova K, Bianucci T, Kimura H, Jülicher F, Honigmann A, Zaburdaev V, Vastenhouw NL. 2021. Transcription organizes euchromatin via microphase separation. Nat Commun 12: 1360. 10.1038/s41467-021-21589-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AG, Smith CW. 2017. Intron retention as a component of regulated gene expression programs. Hum Genet 136: 1043–1057. 10.1007/s00439-017-1791-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I, Schmitt A, Diao Y, Lee AJ, Liu T, Yang D, Tan C, Eom J, Chan M, Chee S, et al. 2019. A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat Genet 51: 1442–1449. 10.1038/s41588-019-0494-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase S, Imai T, Miyauchi-Hara C, Yaguchi K, Nishimoto Y, Fukami SI, Matsuzaki Y, Miyawaki A, Itohara S, Okano H. 2011. Identification of a novel intronic enhancer responsible for the transcriptional regulation of musashi1 in neural stem/progenitor cells. Mol Brain 4: 14. 10.1186/1756-6606-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandekar M, Brandt W, Zhou Y, Dagenais S, Glover TW, Suzuki N, Shimizu R, Yamamoto M, Lim KC, Engel JD. 2007. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development 134: 1703–1712. 10.1242/dev.001297 [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. 2012. Intragenic enhancers act as alternative promoters. Mol Cell 45: 447–458. 10.1016/j.molcel.2011.12.021 [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. 2014. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 9: 2329–2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. 2010. Transcriptional enhancers in animal development and evolution. Curr Biol 20: R754–R763. 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci 100: 189–192. 10.1073/pnas.0136770100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47: W199–W205. 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis R, Karrasch CC, Poulos MG, Kunar B, Redmond D, Duran JG, Badwe CR, Schachterle W, Ginsberg M, Xiang J, et al. 2017. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545: 439–445. 10.1038/nature22326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Liu X, Huang WK, Giusti-Rodríguez P, Cui J, Zhang S, Xu W, Wen Z, Ma S, Rosen JD, et al. 2020. Robust Hi-C maps of enhancer-promoter interactions reveal the function of non-coding genome in neural development and diseases. Mol Cell 79: 521–534.e15. 10.1016/j.molcel.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco-Sola S, Sammeth M, Guigó R, Ribeca P. 2012. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods 9: 1185–1188. 10.1038/nmeth.2221 [DOI] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. The human transcriptome across tissues and individuals. Science 348: 660–665. 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. 2015. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47: 598–606. 10.1038/ng.3286 [DOI] [PubMed] [Google Scholar]
- Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A. 2009. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci 106: 19934–19939. 10.1073/pnas.0900946106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. 2007. Predicting tissue-specific enhancers in the human genome. Genome Res 17: 201–211. 10.1101/gr.5972507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Lluch S, Blanco E, Tilgner H, Curado J, Ruiz-Romero M, Corominas M, Guigó R. 2015. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat Genet 47: 1158–1167. 10.1038/ng.3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervouchine DD, Djebali S, Breschi A, Davis CA, Barja PP, Dobin A, Tanzer A, Lagarde J, Zaleski C, See LH, et al. 2015. Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nat Commun 6: 5903. 10.1038/ncomms6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand E, Cedar H. 2003. Regulation of imprinting: a multi-tiered process. J Cell Biochem 88: 400–407. 10.1002/jcb.10352 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rose AB. 2019. Introns as gene regulators: a brick on the accelerator. Front Genet 9: 672. 10.3389/fgene.2018.00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, et al. 2016. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep 17: 2042–2059. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O. 2017. How introns enhance gene expression. Int J Biochem Cell Biol 91: 145–155. 10.1016/j.biocel.2017.06.016 [DOI] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Revi Genet 15: 272–286. 10.1038/nrg3682 [DOI] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. 2015. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559. 10.1038/nature13994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.