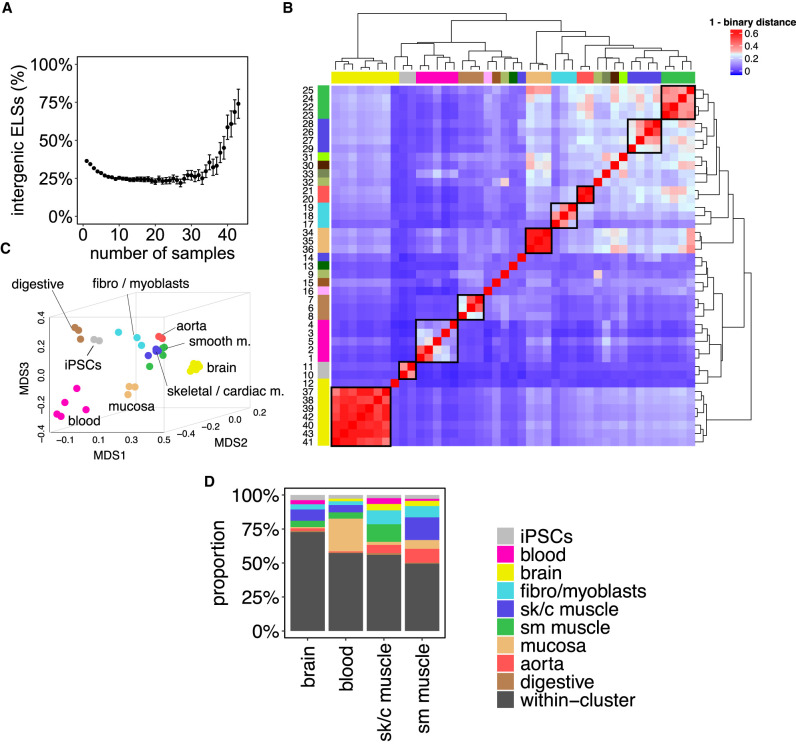
Figure 1.
Active enhancers define tissue identity. (A) Highly shared ELSs are more frequently located in intergenic regions. The scatterplot represents the proportion of intergenic ELSs active in increasing numbers of human adult samples. Error bars represent the 95% confidence interval. (B) Samples’ clustering defined by ELSs’ presence–absence patterns. The heat map depicts the binary distance between any pair of samples, based on the activity of 921,166 distal ELSs (±2 kb from any annotated TSS). The correspondence between samples and numbers is reported in Supplemental Table 1. (C) MDS distribution of human adult samples defined by ELSs’ activity. Analogous representation to Supplemental Figure 1A for the subset of 33 selected adult human samples. (D) Tissue-specific ELSs. The bar plot represents the type of samples found within sets of brain-, blood-, and muscle-specific ELSs. Most tissue-specific ELSs are only active in the samples of the corresponding cluster (“within-cluster”, black), but a few of them may be active in, at most, one outer sample (i.e., a sample that does not belong to the tissue cluster, colored). iPSCs-, fibro-/myoblasts-, digestive-, mucosa-, and aorta-specific ELSs are not represented, because we did not allow outer samples, given their small cluster sizes (see Methods). (sk/c) skeletal/cardiac, (sm) smooth.