Figure 3.
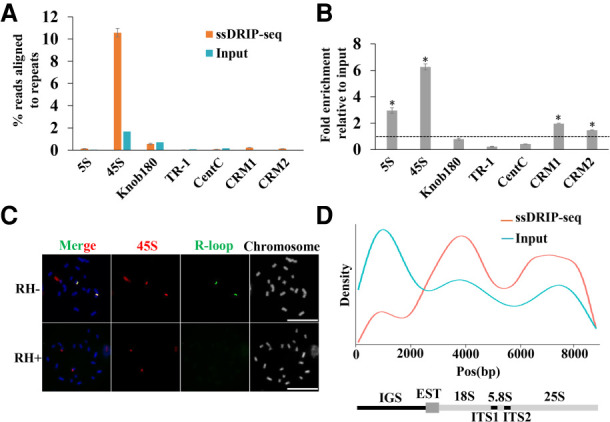
R-loops for tandem repeat sequences. (A,B) Tandem repeat and CRM1/2 consensus sequences were analyzed against the trimmed ssDRIP-seq and input reads using BLAST, independent of mapping to the reference genome, to estimate the abundance of these repetitive sequences in the ssDRIP-seq reads. (A) The percentage of reads corresponding to each tandem repeat sequence and CRM1/2. Data are means ± standard errors (SE) of two biological ssDRIP-seq replicates. (B) The fold enrichment of each tandem repeat and CRM1/2 relative to the amount in input. Data were means ± SE of two biological ssDRIP-seq replicates. Permutation test; asterisks denote the observed value was >90% of the permutation value. (C) R-loops (green) were colocalized to 45S (red) loci on spread chromosomes. RNase H-treatment abolished the R-loop signals on the spread chromosomes. Scale bar = 10 μm. (D) ssDRIP-seq data mapped on the maize 45S rDNA reference sequence shows two enriched regions. Below is the entire 45 rDNA gene locus with intergenic spacer region (IGS).
