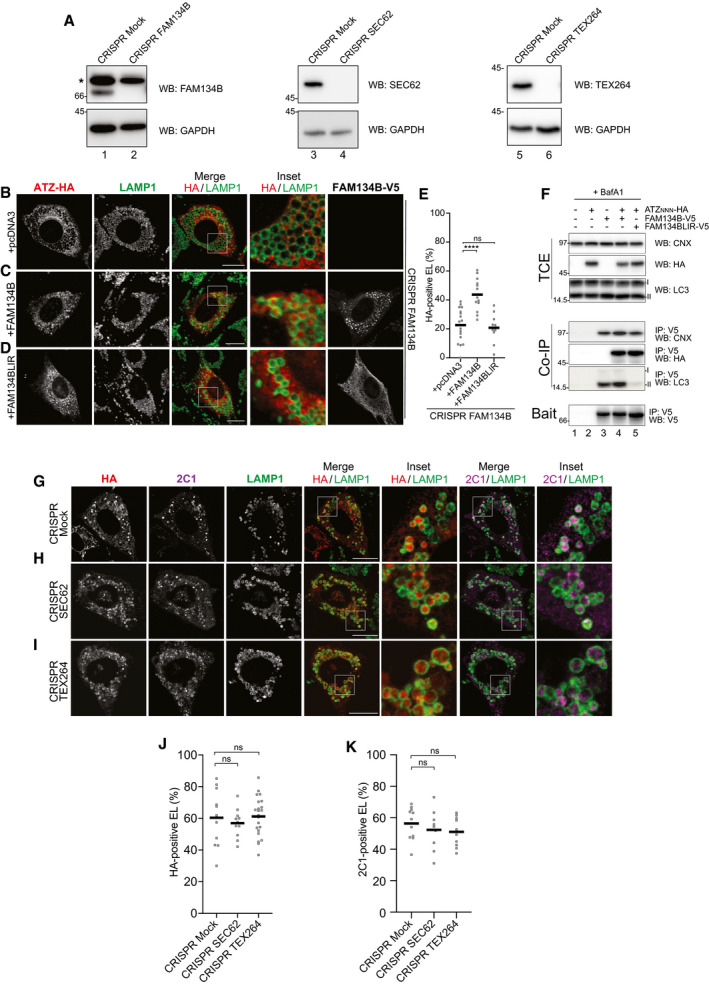
Figure EV1. ER‐phagy receptors involvement in delivery of ATZNNN polymers to endolysosomal compartments.
-
AControl of genetic Fam134B‐, Sec62‐, and Tex264‐KO, respectively. * shows a non‐specific cross‐reaction of the antibody.
- B
-
CSame as (B) in cells back‐transfected with active FAM134B.
-
DSame as (B) in cells back‐transfected with inactive FAM134B (FAM134BLIR), which cannot bind LC3.
-
EQuantification of HA‐positive endolysosomes (B‐D) (mean, n = 17, 13 and 10). One‐way ANOVA and Dunnett’s multiple comparison test, ns P > 0.05, ****P < 0.0001.
-
FHEK293 cells transfected with empty vector (lane 1), ATZ‐HA (2), FAM134B‐V5 (3), FAM134B‐V5 and ATZ‐HA (4), or FAM134BLIR‐V5 and ATZ‐HA (5), treated for 6 h with 100 nM BafA1 and then cross‐linked with DSP before lysis (see Materials and Methods). FAM134BLIR‐V5 interacts with CNX and ATZ‐HA, but not with LC3.
-
G–I(G) Same as (B) in control CRISPR cells and (H) in cells edited with CRISPR/Cas9 to delete Sec62 (Fumagalli et al, 2016) or (I) Tex264.
-
J, K(J) Quantification of (G–I) for HA‐positive endolysosomes, (mean, n = 11, 11, 22) and (K) for 2C1‐positive endolysosomes (mean, n = 11, 9, 11). One‐way ANOVA and Dunnett’s multiple comparison test, ns P > 0.05.
Data information: Scale bars 10 μm.
Source data are available online for this figure.
