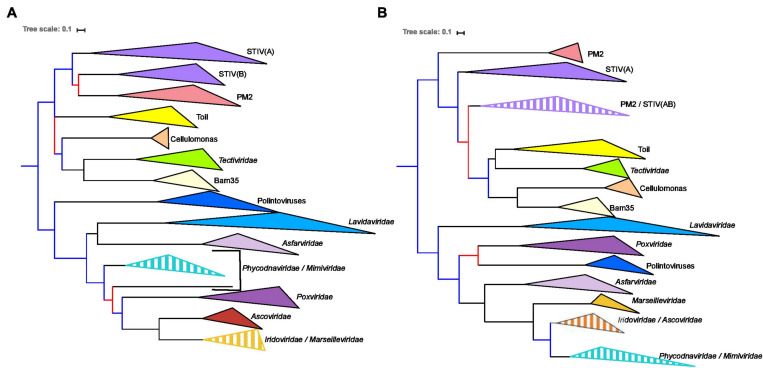
FIGURE 1.
Single-protein trees of the two hallmark proteins of the viruses from the Varidnaviria realm, excluding Adenoviridae, Sphaerolipoviridae. Phylogenetic trees of (A) the major capsid protein (MCP) and (B) packaging ATPase (pATPase). The root of the phylogenetic tree was between the prokaryotic and eukaryotic members. The scale-bar indicates the average number of substitutions per site. The best-fit model for the MCP tree was LG + F + R4, which was chosen according to Bayesian Information Criterion (BIC) and the alignment has 103 sequences with 237 positions. The best-fit model for the pATPase tree was LG + R6, which was chosen according to BIC and the alignment has 103 sequences with 171 positions. More detailed versions of the trees are shown in Supplementary Figures 4, 5. Branches in black indicate both classical bootstrap and transfer bootstrap expectation (TBE) support values using 1,000 replicates are above 70%. Branches in blue indicate only one of the two support values is above 70% whereas branches in red indicate both support values are below 70%.