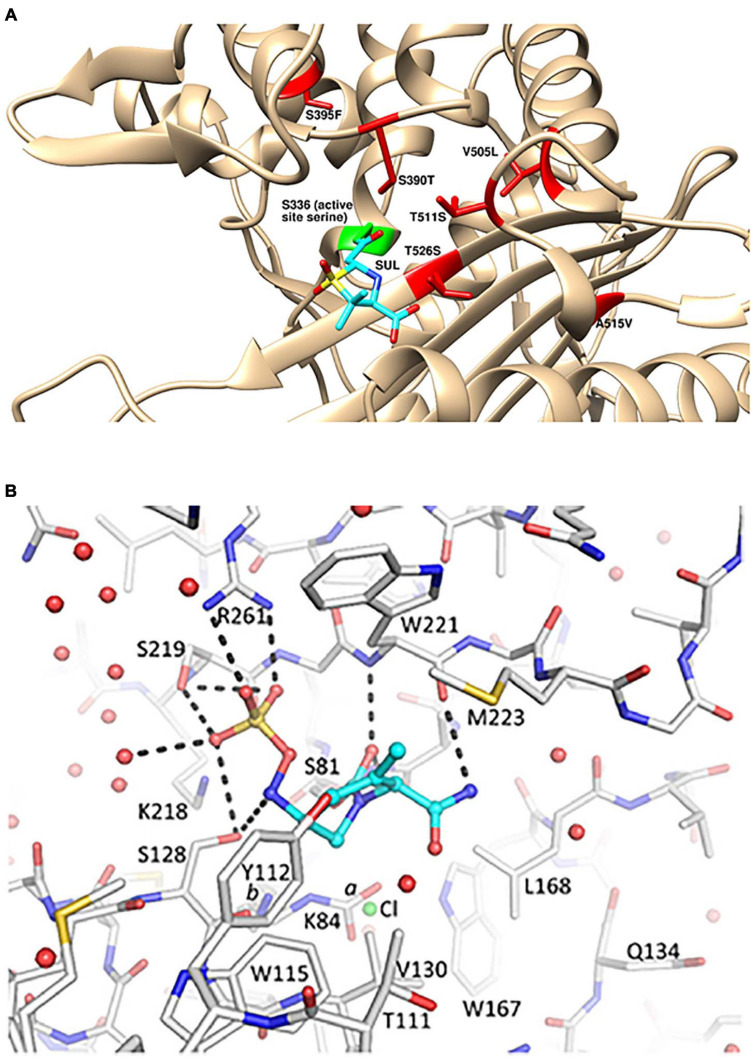
FIGURE 1.
Structural details for the mechanism of action of sulbactam-durlobactam. (A) Computational model of sulbactam bound to the active site of Acinetobacter baumannii PBP3 (PDB: 3UE3). Covalent docking was performed in ICM-Pro version 3.8 and structure visualized in UCSF Chimera version 1.15 (Pettersen et al., 2004), with sulbactam bound to the PBP3 active site serine, shown in green. Sulbactam-resistant mutations are indicated in red and sulbactam is shown in cyan. (B) Durlobactam bound to the active site of OXA24/40 (PDB: 6MPQ). The inhibitor is shown in blue carbon atom stick representation, whereas the protein is depicted in gray carbon stick representation. The K84 side chain was observed in two conformations: one carbamylated and one non-carbamylated [0.6 and 0.4 occupancy conformations labeled a and b, respectively]. A chloride ion with 0.4 occupancy was also refined in the active site (green sphere). Hydrogen bonds between durlobactam and OXA24/40 are shown with dashed lines. Water molecules are depicted as red spheres. Reproduced from Barnes et al. (2019) with permission from the authors.