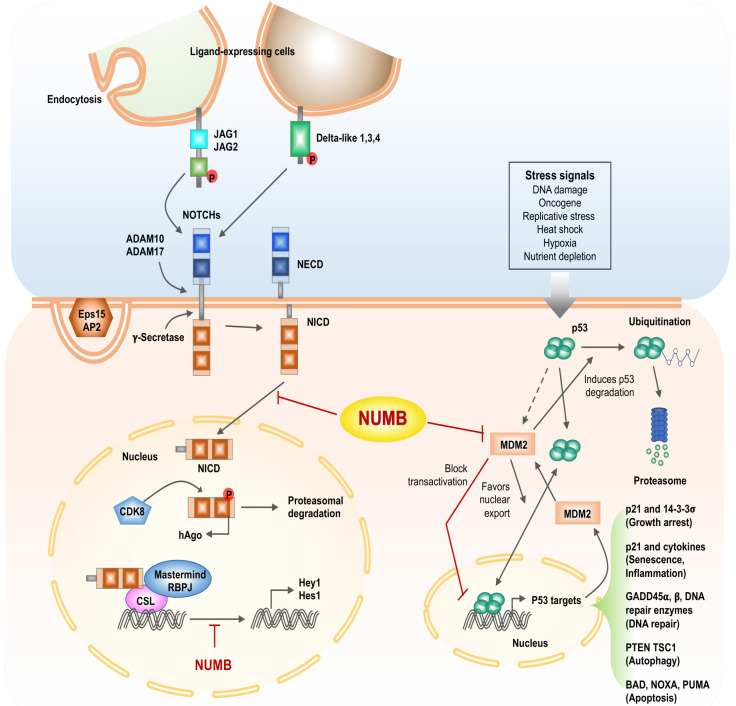
Fig. 3.
Scheme of description for inhibition of the NOTCH signal pathway and regulation of p53 activity by NUMB. Left panel, NUMB interacts with AP2 and Eps15, interfering with the endocytosis of Notch ligands and receptors. The binding of ligands (Delta-like, JAGs) to Notch transmembrane receptors triggers a series of proteolytic cleavages by ADAM proteins (ADAM 10, ADAM 17) and a γ-secretase complex that leads to the release of the Notch intracellular domain (NICD), which is translocated into the nucleus, interacts with the RBPJ (recombining binding protein suppressor of hairless), CSL (CBF1, suppressor of hairless, Lag-1) transcription factor, and the transcriptional activator protein Mastermind to regulate target genes such as Hes1 and Hey1. NUMB can interfere with this transcriptional machinery and inhibit Notch-dependent gene expression. Phosphorylation of NICD by CDK8 leads to hAgo-dependent proteosomal degradation of NICD. NUMB inhibits translocation of NCID into the nucleus. Right panel, p53 activity is driven by several stress-related signals. p53-dependent transcription of MDM2 increases p53 degradation. MDM2 suppresses p53 activity by blocking p53 transcriptional activity and favoring p53 nuclear export. Transcription of p53 target genes promotes p53 tumor-suppressor properties.