Abstract
In this proof-of-concept work, we have developed a 3D-CNN architecture that is guided by the tumor mask for classifying several patient-outcomes in breast cancer from the respective 3D dynamic contrast-enhanced MRI (DCE-MRI) images. The tumor masks on DCE-MRI images were generated using pre- and post-contrast images and validated by experienced radiologists. We show that our proposed mask-guided classification has a higher accuracy than that from either the full image without tumor masks (including background) or the masked voxels only. We have used two patient outcomes for this study: (1) recurrence of cancer after 5 years of imaging and (2) HER2 status, for comparing accuracies of different models. By looking at the activation maps, we conclude that an image-based prediction model using 3D-CNN could be improved by even a conservatively generated mask, rather than overly trusting an unguided, blind 3D-CNN. A blind CNN may classify accurately enough, while its attention may really be focused on a remote region within 3D images. On the other hand, only using a conservatively segmented region may not be as good for classification as using full images but forcing the model’s attention toward the known regions of interest.
Keywords: Deep learning, Breast cancer outcome classification, DCE-MRI, Mask-guided convolutional neural net
Introduction
Breast cancer is the most common non-skin cancer in adult women worldwide, and the second leading cause of cancer deaths in women [1]. Among various medical imaging tools for breast cancer, dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is highly sensitive for the detection and evaluation of breast cancer in all breast cancers. DCE-MRI is highly sensitive for detecting breast cancer, particularly in high-risk populations with a higher sensitivity than mammography and ultrasound [2, 3]. Since DCE-MR images contain information on tumor perfusion and heterogeneity, they may provide features that are not visually apparent but highly correlated with tumor phenotype and prognosis. The entire field of radiomics is based on the ability to find associations between imaging data and outcomes, though very little has been published on comparison of the methods revealing those associations.
Classification models based on convolutional neural network (CNN) architectures have shown potential solutions for revealing the hidden relationship between breast tumor prognosis and medical images [4, 5]. Commonly used 2D-CNN architectures only use the spatial information in an axial plane without evaluating the pixel correlation in the coronal plane and sagittal plane. It has been shown that 3D-CNN outperforms 2D-CNN methods in many other studies [6, 7]. However, most of these studies presented 3D-CNN as a black box and did not visually explain where the model’s attention lies in those images.
In reality, most tumor datasets contain invasive and non-invasive tumors within the same field of view. For example, 80% of breast cancers are categorized as invasive breast cancer, which tends to grow out of the originated lesion. The current methods are not explicitly designed for classifying the tumor outcome within invasive and non-invasive tumor datasets. Some of those studies cropped only the tumor volume out of the entire image set, regardless of the correlation between the tumor and surrounding tissue [5]. Other studies applied 3D-CNN on classifying disease outcomes for volumetrically diffusive conditions like Alzheimer’s disease [7, 8]. Therefore, classifying disease outcomes using tumors that contain both invasive and non-invasive tumor tissues still lacks extensive study.
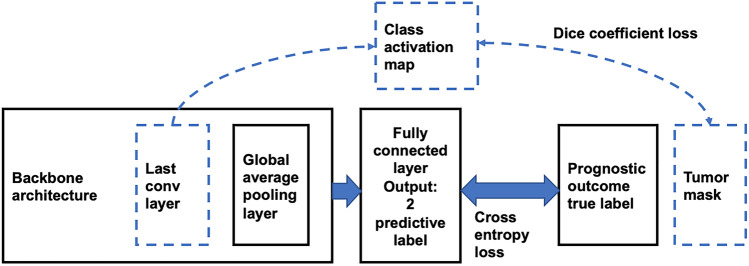
We have previously established the ability of radiomics combined with conventional machine learning classification models on MR and positron emission tomography (PET) images acquired at the time of diagnosis to predict various breast cancer outcomes, including survival [8]. In this paper, we propose to use a new mask-guided 3D-CNN architecture for tumor-outcome prediction from breast DCE-MRI, which improves the classification performance and, more importantly, guides the 3D-CNN’s attention toward the breast tumor. We generated tumor masks first using the difference between pre- and post-contrast DCE-MRI images, which are then validated by two experienced radiologists. We consider the attention of the 3D-CNN as the highlighted region on the activation map [9]. Our mask-guided 3D-CNN architecture is a classification method based on maximizing the dice coefficient [11] between the mask and highlighted attention of the 3D-CNN. The dice coefficient quantifies the degree of overlap. In this way, we were able to guide the attention of 3D-CNN from outside the breast to the region of interest, which is close to the tumor. In this study, we found that our mask-guided 3D-CNN approach outperformed the unguided 3D-CNN approach and tumor-only approach in classifying various breast cancer outcomes, including cancer recurrences and human epidermal growth factor receptor 2 (HER2) status. We present here results for only these two outcomes as the dataset had class balance for them, and consequently, the results are more reliable. Additionally, the survival is an important prognostic parameter, and HER2 has a strong possibility of correlating with image features. These rationales also played a key role in our choice for outcomes to be correlated with. The tumor-only method uses only the segmented tumor volume.
Proposed Method
Backbone Architecture
We chose a modified 3D-VGGNet [12, 13] for binary classification studies as a backbone architecture in this study. To visualize the highlighted region for breast cancer by 3D-CNN, we replaced typical fully connected layers with global average pooling layers, as shown in Fig. 1. Notably, the input images pass through 4 blocks constructed by 3D convolutional layers and 3D max-pooling layers. The architecture was followed by a global average pooling layer, batch normalization layer [14], dropout layer [15], and the softmax output layer. For tuning of the 3D-CNN, we used adaptive moment estimation (Adam) [16] optimizer with a 0.000025 learning rate. We trained 250 epochs with a batch size of 10.
Fig. 1.
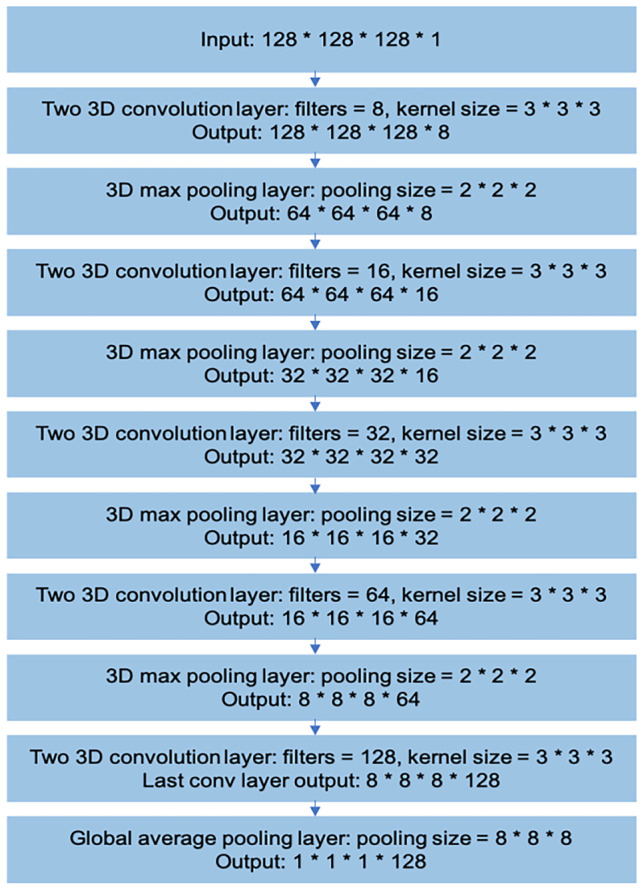
Modified 3D VGGNet backbone architecture
Visualization
In 3D-CNN medical image analysis studies, one of the major problems is the lack of transparency inside the neural network. In this study, we used the class activation map (CAM) with global average pooling (GAP) [10] to visualize discriminative image regions by 3D-CNN. A global average pooling layer and a fully connected layer were added at the end of the last 3D convolutional layer. This design allowed the projection of class label weights of the output layer onto the activation maps in the convolutional layer. Thus, it enabled us to identify the critical image region by projecting back the weights of the output layer onto the 3D convolutional feature maps.
Loss Function
In order to test if the mask-guided mechanism is able to help increase the model classification performance, we investigated the visualization explanation and classification performance of the mask-guided 3D-CNN by comparing three different models. The summary of these three models is shown in Fig. 2 and Table 1. All of these models that classified breast cancer prognostic outcomes were based on the 3D-VGGNet architecture with the modified backbone mentioned above. We adjusted the dimensions of input images for different models. In the no-mask model (Table 1), we took the whole 3D DCE-MR volume as input. Additionally, we selected the binary cross-entropy function calculated by the probabilities output and the ground truth labels as our loss function (function 1). For the mask-only model, we used the 3D tumor boundary boxes as input. The mask-only model used the same loss function as the no-mask model. Finally, in our proposed mask-guided model, we again used 3D DCE-MR volumes as input. To guide the attention of the 3D-CNN to the tissue highlighted by contrast-enhanced tissue, we created a loss function that is able to minimize the cross-entropy and the area difference between the class-activation-map and the tumor mask generated by contrast-enhanced breast tissue at the same time. The loss function is shown below (function 2). We set our hyperparameter, λ, to equal to 10, as it appears to adequately balance the two terms in function 2.
Fig. 2.
The schema for our proposed architecture. Models 1 and 2 are without the dashed lines. Model 3 is the one model with the dashed lines
Table 1.
Input, output, and loss function difference of three models
| Input | Output layer | Loss function | |
|---|---|---|---|
| Model 1: not using the segmented mask | 3D MRI volume | The output of a fully connected layer |
Cross-entropy between the true label and predictive label (function 1, see below) |
| Model 2: using only the segmented mask | Only voxels within the boundary box of tumor mask | The output of a fully connected layer |
Cross-entropy between the true label and predictive label (function 1) |
| Model 3: our novel mask-guided model | 3D MR volume | The output of a fully connected layer and last conv output |
1: Cross-entropy between a true label and predictive label 2: Dice coefficient between tumor mask and 3D CNN activation map (function 2) |
Function 1: CE (L, P) = −Σ Li log(Pi)
DiceCoef (mask1, mask2) = 2 × |mask1 ∩ mask2|/(|mask1| +|mask2|)
Function 2: loss = CE (label, prediction) − λ × DiceCoef (class activation map, mask)
CE refers to cross-entropy, i refers to different images, L refers to label, P refers to prediction
Experiment and Results
Dataset
We evaluated the classification performance of the mask-guided model on 3D DCE-MR breast images, which consists of a collection of 115 pre-contrast (T0), early post-contrast (T1), and late post-contrast (T2) DCE-MR images. In this study, we predicted recurrence (pos/neg = 35/80) and HER2 status (pos/neg = 30/79) as our target labels. Retrospective data that we used were acquired with 1.5 T (SIGNA, GE Healthcare) or 3.0 T (Magnetom, Siemens Healthineers), with contrast agent gadopentetate dimeglumine (0.1 mmol/kg). The acquisition details for the datasets used in this study were described previously. [17] All imaging study was performed at the University of California, San Francisco, under Institutional Review Board–approved protocol. The retrospective data analysis was approved by the Institutional Review Board.
To generate the 3D tumor volume mask, we subtracted the background pre-contrast image set (T0) from the early post-contrast image set (T1) to obtain the subtracted contrast-only image set. We set the contrast threshold at 70%, only considering contrast greater than 70% contrast enhancement threshold based on the past experience with this dataset that is standard in MRI [9]. We did not use T2 image as the signal were noisy and outside the tumor. We segmented breast tumor tissue using a fuzzy C-means algorithm and created a tumor volume mask to guide this model. Due to the peak contrast of breast tissue appearing at early post-contrast (T1), we chose T1 as the training input image for our analysis. As input to our mask-only model, the tumor volume was manually segmented from DCE-MRI images, confirmed, and validated under the guidance of experienced breast radiologists trained in breast imaging [17]. One of our co-authors (M.B.) performed the validation of the mask under supervision of a senior radiologist (D.W.). Both of them have subspeciality training in breast imaging. All of radiologists (M.B., D.W., and B.J.) reviewed the class activation maps included in this paper.
A total of 115 subjects were randomly shuffled and divided into training and validation datasets by five-fold cross-validation. We then developed a 3D data augmentation method and generated 92 (four-folds) × 34 = 3128 training images. And we used 23 original images without augmentation for testing. Specifically, we first rotated the original 3D images four times on the horizontal surface. Because there are six faces in a 3D cube, we were able to generate 4 × 6 = 24 augmented images. Second, we shifted the original 3D image along with the directions of eight cube angles and filled with a background pixel value. We shifted 10 pixels along x-, y-, z-axes as follows ([[10, 10, 10], [10, 10, −10], [10, −10, 10], [10, −10, −10], [−10, −10, −10], [−10, −10, 10], [−10, 10, −10], [−10, 10, 10]]). We generated eight new images in this process. Finally, we rotated the original 3D original image 10 degrees clockwise and anticlockwise on the horizontal surface to produce two more images. Within these three processes, we augmented each image 24 + 8 + 2 = 34 times.
Visualization Results
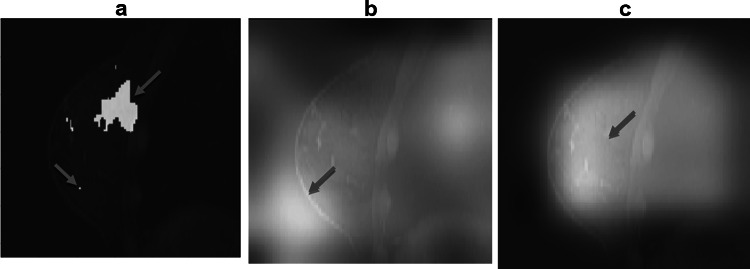
After we trained our model to predict the prognostic outcome label, we visualized our attentions of no-mask and mask-guided model on the original image. The heatmap images of activation in Fig. 3b, c show the attentions of the no-mask model and mask-guided model. This result indicated that the mask-guided technique was able to guide the attention of the 3D convolutional neural network inside the breast (3c) instead of the non-relevant region such as lung or region outside of the breast tissue as for the non-mask-guided one (3b). Since tumor segmentation (3a) used a very conservative approach, we conjecture that the region of attention in (3c) has the essential image features relevant to our target classification labels.
Fig. 3.
a Shows the tumor mask, extra pixels, e.g., one in the left lower quadrant, are present because the way segmentation is performed from the difference image between contrast-enhanced and no-contrast MRI, b visualizes that the class activation map of no-mask model 2 is off the breast tissue, and c visualizes that the class activation map of mask-guided model 3 finds the regions of interest inside the breast but outside the mask
Accuracy of Results
We used five-fold cross-validation to evaluate the classification using the mask-guided technique. We compared the predictive performance of three 3D-CNN models on two prognostic outcomes. The results are shown in Tables 2 and 3. The prediction was measured by the area under the receiver operating characteristic curve (AUC) and classification accuracy (ACC). Five-fold cross-validation was conducted to calculate the standard deviations.
Table 2.
Classification performance in predicting HER2 status. Mask-guided model 3 is consistently the best
| ACC (training) | ACC (validation) | AUC (training) | AUC (validation) | |
|---|---|---|---|---|
| Model 1 |
0.776 (SD 0.034) |
0.749 (SD 0.071) |
0.801 (SD 0.046) |
0.778 (SD 0.065) |
| Model 2 |
0.766 (SD 0.045) |
0.638 (SD 0.157) |
0.790 (SD 0.082) |
0.695 (SD 0.128) |
| Model 3 |
0.781 (SD 0.046) |
0.762 (SD 0.066) |
0.812 (SD 0.051) |
0.781 (SD 0.036) |
Table 3.
Classification performance of three models on predicting recurrence in 5 years. Model 1 looks into the only masked area (Fig. 3b). Model 2 looks into the whole image without the mask information. Model 3 is also over the entire image but mask-guided. As in Table 2, mask-guided model 3 classifies outcomes slightly and consistently better than the other two
| ACC (training) | ACC (validation) | AUC (training) | AUC (validation) | |
|---|---|---|---|---|
| Model 1 |
0.798 (SD 0.038) |
0.758 (SD 0.065) |
0.822 (SD 0.046) |
0.798 (SD 0.077) |
| Model 2 |
0.783 (SD 0.027) |
0.672 (SD 0.169) |
0.810 (SD 0.043) |
0.705 (SD 0.184) |
| Model 3 |
0.823 (SD 0.036) |
0.796 (SD 0.076) |
0.844 (SD 0.041) |
0.828 (SD 0.068) |
Discussion
Our results in Tables 2 and 3 show that forcing the mask-guided model 3 to pay attention toward the tumor volume, rather than the surrounding tissue, improves the prediction results. Furthermore, our use of 3D augmentation for creating extensive training data set reduced the potential of over-fitting even though our initial dataset was small (n = 115) in this study.
For the visualization of results, the activation map in Fig. 3b showed that the no-mask model 1 directed the attention to the edge between the breast and the background, which indicated that the no-mask mode 1 classified the tumor with high accuracy based on unreliable image information from outside of the tumor. This is a known bizarre unexplained property of deep learning. It is possible that there is some associated noise feature in the image that is highly correlated with the labels without necessarily having any causal relationship. This may be due to the small size of the training dataset (n = 115). Another novelty of the present study is that, unlike other prognostic outcome classification studies [15, 18], we used full 3D images for training. This high dimensionality of training images may have directed the model’s attention toward the region of interest. As the original manually drawn mask may have been done conservatively, the neural network found a larger region of interest within the breast for tumor classification (Fig. 3c), which may be correlated to the breast cancer subtype characteristics. By “larger region,” we mean outside the segmented mask. The resulting performance has improved both accuracies (ACC), and prediction (AUC) compared to the other model 2 and model 3 (Table 3).
Results shown in Table 3 demonstrated that the mask-guided model outperformed the other two models in predicting 5-year disease-free survival in our cohort of breast cancer patients. Model 3 (mask-guided) was better than model 1 (no-mask) because the mask-guided model used the voxels within the tumor that were relevant to the treatment response. Compared to model 2 (mask-only), model 3 (mask-guided) classified the outcome not only based on the tumor tissue within the boundary box but also assessed the surrounding activated stroma that may also relate to DFS [19]. This type of guidance of CNN is not attempted before, to the extent of our knowledge.
In this study, a contrast difference between the early post-contrast images at T1 and pre-contrast at T0 images is used to generate the breast tumor masks. This method was able to highlight the suspected tumor tissue. However, this segmentation approach occasionally created irregular tumor masks or multiple small tumor chunks for some patients. Due to the varying size of the discontinuous tumor chunks within one patient and between patients, the existing CNN could provide only two types of models: no-mask model or mask-only model. No-mask model did not use the tumor mask generated from DCE-MRI. On the other hand, the mask-only model may introduce noise into the dataset before training, because of a necessary image-resizing step. In such circumstances, it is advantageous to use the mask-guided technique on 3D images that do not require tumor resizing but uses all critical information on the DCE-MRI image for accurate classification and prediction.
Conclusion
In this paper, our novel 3D-CNN architecture has used the tumor mask as a guide to direct 3D-CNN’s attention for classification. Our mask-guided model was designed to focus on tumors possibly present in different regions over a breast. We observed that the mask-guided model was able to focus its attention at an unknown region within the breast tumor that was different from the mask, possibly indicating image features relevant to the target prognostic tumor outcomes. This technique also helped to increase the accuracy of classification performance on outcomes over the DCE-MRI dataset that we tested. We expect this result will motivate the community on follow-up larger-scale projects, and if our results are replicated, then utilize our proposed mask-guided method to develop novel image bio-markers.
A limitation of our work was to use a too conservative tumor segmentation algorithm (for model 2) that may have excluded some tumor tissues. It is worth comparing model 3 against model 2 with a more inclusive segmentation approach. However, the fact that even a conservative segmentation algorithm may be used to guide patient-outcome prediction is a novel contribution of our work worth further pursuing with a larger dataset than those that were used in this study. Small dataset raises obvious question on stability of our results. An alternative to deep learning-based approach is to use machine learning over radiomics or image features for prediction of patient outcomes [19, 20], which should be compared against our proposed method (model 3).
Funding
This study was supported in part by the National Institute of Biomedical Imaging and Bioengineering grant R01EB026331. We thank anonymous reviewers in improving the article and for suggestions on future work.
Declarations
Conflict of Interest
There is no known conflict of interest from all authors, except for Mitra, who is a co-founder of SolvingDynamics, Inc. SolvingDynamics was not related to this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1. DeSantis, Carol E., et al.: Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA: a cancer journal for clinicians 66.1 (2016): 31–42. [DOI] [PubMed]
- 2. Nie, Ke, et al.: Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Academic radiology 15.12 (2008): 1513–1525. [DOI] [PMC free article] [PubMed]
- 3.Warner E, et al. Comparison of breast magnetic resonance imaging, mammography, and ultrasound for surveillance of women at high risk for hereditary breast cancer. Journal of Clinical Oncology. 2001;19(15):3524–3531. doi: 10.1200/JCO.2001.19.15.3524. [DOI] [PubMed] [Google Scholar]
- 4.O'Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21(2):249–257. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Couture, H.D., Williams, L.A., Geradts, J. et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. npj Breast Cancer 4, 30 (2018). 10.1038/s41523-018-0079-1. [DOI] [PMC free article] [PubMed]
- 6.Li J, Fan M, Zhang J, Li L.: Discriminating between benign and malignant breast tumors using 3D convolutional neural network in dynamic contrast enhanced-MR images. In Medical Imaging 2017: Imaging Informatics for Healthcare, Research, and Applications 2017 Mar 13 (Vol. 10138, p. 1013808). International Society for Optics and Photonics.
- 7.Hosseini-Asl E, Gimel'farb G, El-Baz A.: Alzheimer's disease diagnostics by a deeply supervised adaptable 3D convolutional network. arXiv preprint arXiv1607.00556. 2016 Jul 2. [DOI] [PubMed]
- 8.Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, Lituiev D, Copeland TP, Aboian MS, Mari Aparici C, Behr SC. A Deep learning model to predict a diagnosis of alzheimer disease by using 18F-FDG PET of the brain. Radiology. 2018;290(2):456–464. doi: 10.1148/radiol.2018180958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang SY, Franc BL, Harnish RJ, Liu G, Mitra D, Copeland TP, Arasu VA, Kornak J, Jones EF, Behr SC, Hylton NM. Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ breast cancer. 2018;4(1):24. doi: 10.1038/s41523-018-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A.: Learning deep features for discriminative localization. InProceedings of the IEEE conference on computer vision and pattern recognition 2016 (pp. 2921-2929).
- 11.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. doi: 10.2307/1932409. [DOI] [Google Scholar]
- 12.Korolev S, Safiullin A, Belyaev M, Dodonova Y.: Residual and plain convolutional neural networks for 3D brain MRI classification. In2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 2017 Apr 18 (pp. 835–838). IEEE.
- 13.Yang C, Rangarajan A, Ranka S.: Visual explanations from deep 3D convolutional neural networks for Alzheimer's disease classification. arXiv preprint arXiv1803.02544. 2018 Mar 7. [PMC free article] [PubMed]
- 14.Ioffe, Sergey, and Christian Szegedy.: Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv1502.03167. 2015.
- 15.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. The Journal of Machine Learning Research. 2014;15(1):1929–1958. [Google Scholar]
- 16.Kingma DP, Ba J. Adam.: A method for stochastic optimization. arXiv preprint. arXiv1412.6980 2014 Dec 22.
- 17.Bolouri MS, Elias SG, Wisner DJ, Behr SC, Hawkins RA, Suzuki SA, Banfield KS, Joe BN, Hylton NM. Triple-negative and non-triple-negative invasive breast cancer: association between MR and fluorine 18 fluorodeoxyglucose PET Imaging. Radiology. 2013;269(2):354–361. doi: 10.1148/radiol.13130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones EF, Sinha SP, Newitt DC, Klifa C, Kornak J, Park CC, Hylton NM.: MRI enhancement in stromal tissue surrounding breast tumors: association with recurrence free survival following neoadjuvant chemotherapy. PloS one. 2013;8(5). [DOI] [PMC free article] [PubMed]
- 19.Jiang Z, Song L, Lu H, Yin J. The potential use of DCE-MRI texture analysis to predict HER2 2+ status. Front Oncol. 2019;9:242. doi: 10.3389/fonc.2019.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Imbriaco M, Cuocolo R: Does texture analysis of MR images of breast tumors help predict response to treatment? Radiology. 2018; 286(2). 10.1148/radiol.2017172454. [DOI] [PubMed]