Figure 3.
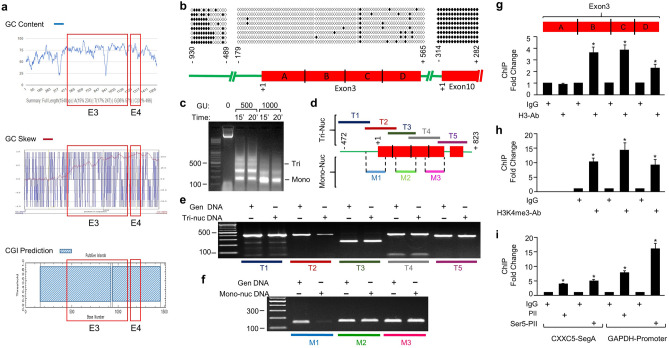
Feature of the CXXC5 region encompassing the promoter of CXXC5. (a) In silico analyses of the nucleotide sequence of the region for G-C content, asymmetric GC distribution, GC skew (GC skew), and the presence of a CpG island (CGI). (b) The methylation state of the CXXC5 promoter region (-930 to -489 and -179 through the Exon3; + 1 indicates the beginning of Exon3) together with the 3’-end of Intron9 and Exon10 (− 195 through + 330; + 1 marks the beginning of Exon10) as controls was examined with bisulfite sequencing. Isolated genomic DNA of MCF7 cells was subjected to bisulfite reaction for the conversion of unmethylated cytosine residues to uracil followed by bisulfite PCR. PCR amplicons produced with bisulfite primers were cloned and sequenced. Aligned sequences to the corresponding CXXC5 regions were depicted as a lollipop distribution. Filled circles indicate methylated and empty circles denote unmethylated CpG dinucleotides. (c) Nucleosome occupancy at the CXXC5 promoter elements was assessed with Micrococcal Nuclease (MNase) assay. MCF7 cells were fixed, permeabilized, and treated without (0) or with 500 or 1000 gel units (GU) of MNase for 15 or 20 min at 37 °C for chromatin digestion. Isolated DNA was analyzed with agarose gel electrophoresis. (d) Schematics of regions subjected to PCR. Isolated DNA fragments corresponding to (e) tri-nucleosomal (T1-5) and (f) mono-nucleosomal (M1-3) DNA were subjected to PCR using region-specific primer pairs. (g–i) ChIP analysis of Exon3. Following chromatin digestion of MCF7 cells by the use of MNase were subjected to ChIP using species-specific IgG, an antibody specific to total H3 (g) or H3K4me3 (h). Isolated DNA following precipitation with Protein A/G conjugated magnetic beads was subjected to qPCR using region-specific primer sets. Asterisk (*) denotes significant changes depicted as fold change compared to IgG. (i) MCF7 cells fixed, lysed, and sonicated were subjected to ChIP using IgG, PolII, or ser5-phosphorylated PolII antibody followed by precipitation with Protein A/G conjugated magnetic beads. DNA samples were then used for qPCR with primer sets specific to Segment A or the promoter of GAPDH as control. Shown are the mean ± SE of three independent experiments performed in triplicate. Significant differences depicted with an asterisk (*) are shown as fold change compared to IgG.