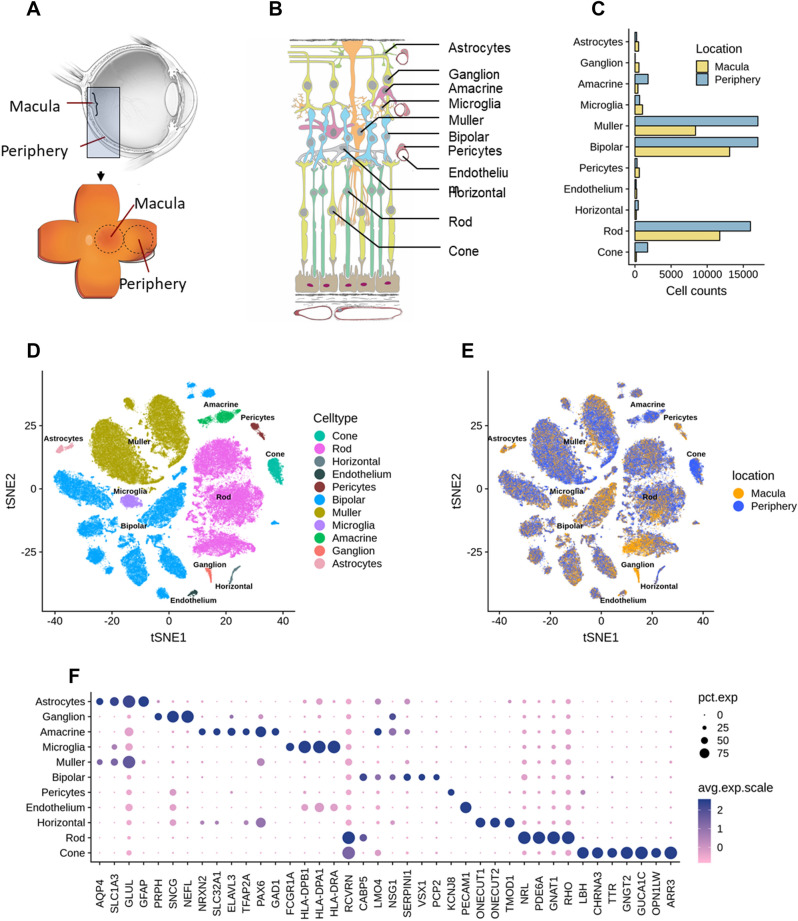
Figure 1.
Summary of single-cell analysis from human retina. (A) Schematic cross-section of human eye (top) showing the retina lining the interior surface. The macula contains the fovea and is responsible for sharp vision. The periphery is responsible for detecting light and motion. Schematic of dissected tissue (bottom) shows retina adjoined to support tissues, flattened with relaxing cuts. Areas 8 mm in diameter were excised for RNA sequencing. (B) Layers of human retina and supporting tissues showing 11 assayed cell types. Five neuronal classes are photoreceptors, bipolar cells, ganglion cells, horizontal and amacrine cells. Cone photoreceptors are sensitive to color and bright light. Rod photoreceptors are sensitive to low light. Ganglion cells transmit information to the brain. Horizontal cells and amacrine cells modulate signal from photoreceptors and bipolar cells, respectively. Müller glia span the retina and are involved in neurotransmission, fluid balance, and wound repair. Also depicted are microglia (with phagocytic and immune activity), astrocytes (regulation of metabolism and blood brain barrier, synaptogenesis, neurotransmission), vascular endothelium (vascular tone and blood flow; coagulation and fibrinolysis; immune response, inflammation and angiogenesis) and pericytes (integrity of endothelial cells, trans-regulation of vascular tone, stem cells). The retinal layers include: NFL, nerve fiber layer; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; IS, OS, inner segments and outer segments of photoreceptors. Below the rods and cones are (from upper to lower) retinal pigment epithelium, Bruch’s membrane, and choriocapillaris, which are shown for completeness and were not assayed. (C) Bar plots showing proportions of counts of cells in each identified cell types from the scRNA-seq data across the two retina regions. Note that the counts of cells in each cell type do not reflect cell type composition in the tissue. (D) Visualization of scRNA-seq clusters from combined macula and periphery using t-SNE. Cells are colored by cell types. (E) Visualization of scRNA-seq clusters using t-SNE. Cells are colored by region of origin-macular or periphery. Note clusters are represented by both macula and peripheral regions. (F) Dot plots showing expression pattern of known gene markers across cell types (Supplementary data 1).