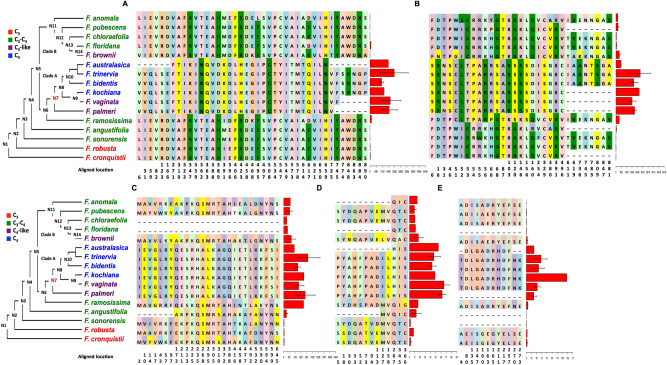
Figure 2.
Modifications in genes in C4 pathway in predicted protein sequences and transcript abundances mapped to the Flaveria phylogeny. The predicted amino acid changes and the transcript abundance (FPKM) of the genes encoding the enzymes in C4 pathway are shown. Only the amino acid residues predicted to be different between C3 and C4 species are superimposed on the recent published phylogeny of Flaveria. The colors of amino acid residues have no meaning and are only for visualization purposes. Numbers below the amino acids indicate the location sites in the multiple sequence alignments. FPKM values with standard errors are shown to the right of the amino acid changes as red bars. (A) phosphoenolpyruvate carboxylase (PEPC); (B) pyruvate orthophosphate dikinase (PPDK); (C) NADP-malic enzyme (NADP-ME); (D) pyruvate orthophosphate dikinase regulatory protein (PPDK-RP); (E) phosphoenolpyruvate protein kinase A (PPCKA). Protein sequences from UniprotKP are: F. trinervia PEPC, P30694; F. bidentis PPDK, Q39735; F. brownii PPDK, Q39734; and F. trinervia PPDK, P22221. Sequence alignments are available in Additional file 2.