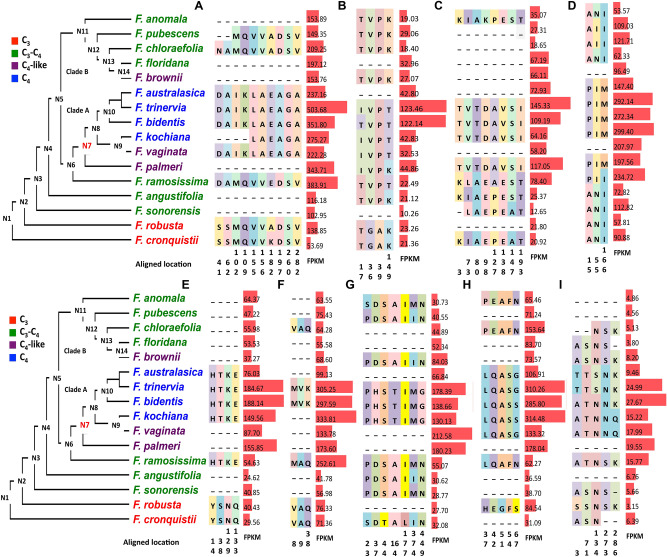
Figure 3.
Modifications in the predicted amino acid sequences of proteins involved in cyclic electron transport and transcript abundances of the cognate transcripts mapped to the Flaveria phylogeny. Changes in predicted amino acid sequence in proteins involved in cyclic electron transport chain and abundances (FPKM) of their cognate transcripts in C4 and C3 Flaveria species are shown. Only the amino acid residues predicted to be different between C3 and C4 species are superimposed recent published phylogeny of Flaveria. The marked colors of amino acid residues have no meaning and are only for visualization purposes. Numbers below the amino acids indicate the location site in the multiple sequence alignments. FPKM values with standard errors are represented to the right of the amino acid changes as red bars. (A) Protein gradient regulation 5 like protein (PGR5-like); (B) NADH dehydrogenase-like (Ndh) L2 subunit (Ndh L2); (C) NdhV; (D) Ndh16; (E) NdhU; (F) NdhM; (G) Ndh48; (H) NdhB4; (I) chlororespiration reduction 1 (CRR1). The sequence alignments are available in Additional file 2.