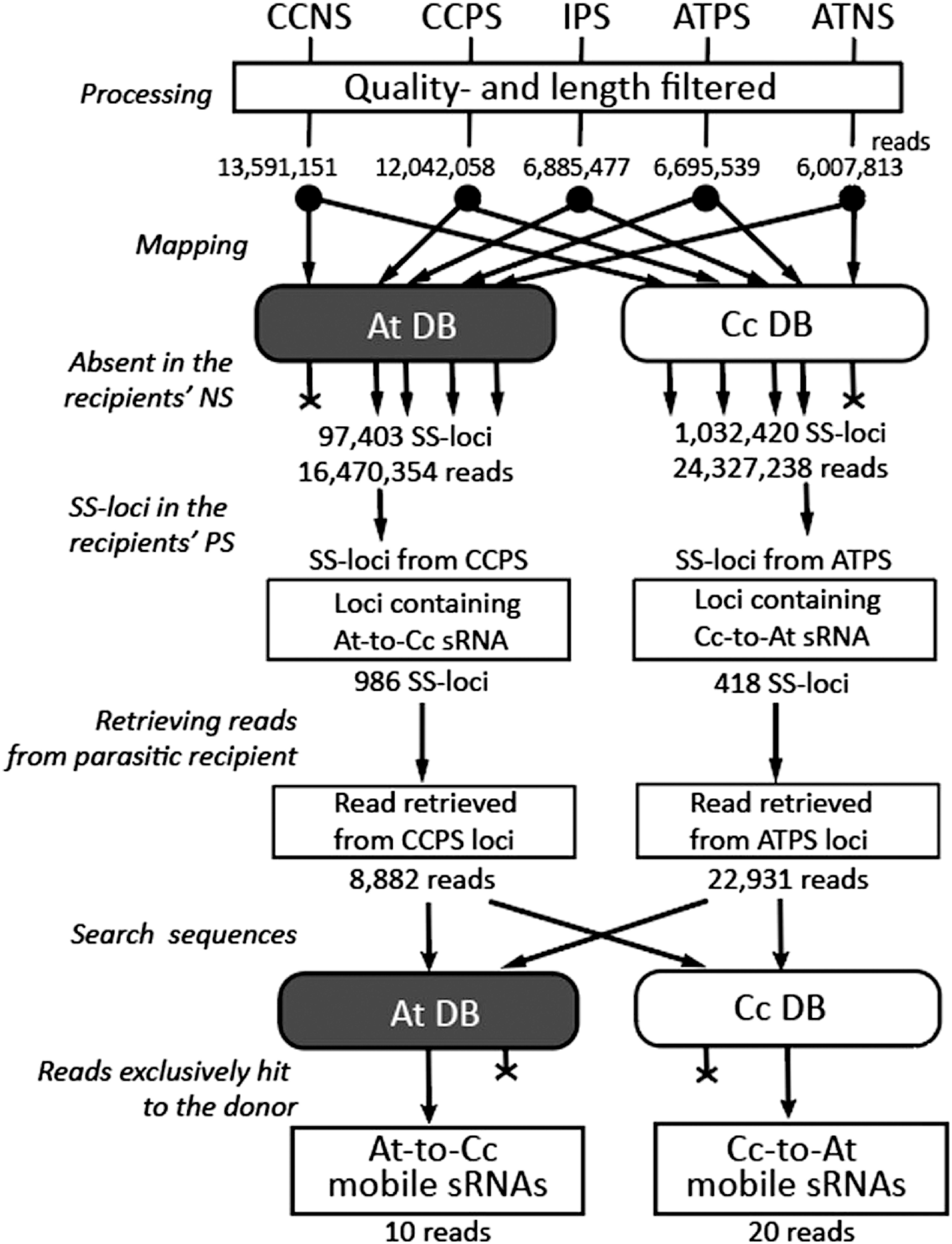
Figure 3. Flowchart of mobile sRNA screening in the Cuscuta campestris-Arabidopsis thaliana parasitic complex. sRNA-Seq reads obtained from the indicated tissues (Reads) were quality- and length-filtered (Processing), and mapped to the donor and recipient genome databases (Mapping). ShortStack-loci (SS-loci) absent in the nonparasitic recipient were selected (Absent in the recipients’ PS), and reads were retrieved from the parasitic recipient libraries (SS-loci in the recipients’ PS, Retrieve reads from parasitic recipient). The retrieved reads were searched in both donor and recipient genomes (Search sequences) and the reads, which were exclusive hits of the donor genome, were regarded as plant-to-plant mobile sRNAs (Reads exclusively hit to the donor). Cc, Cuscuta campestris; At, Arabidopsis thaliana, DB, genome sequence database. Numbers indicate numbers of reads or loci that remained after the procedures just before.