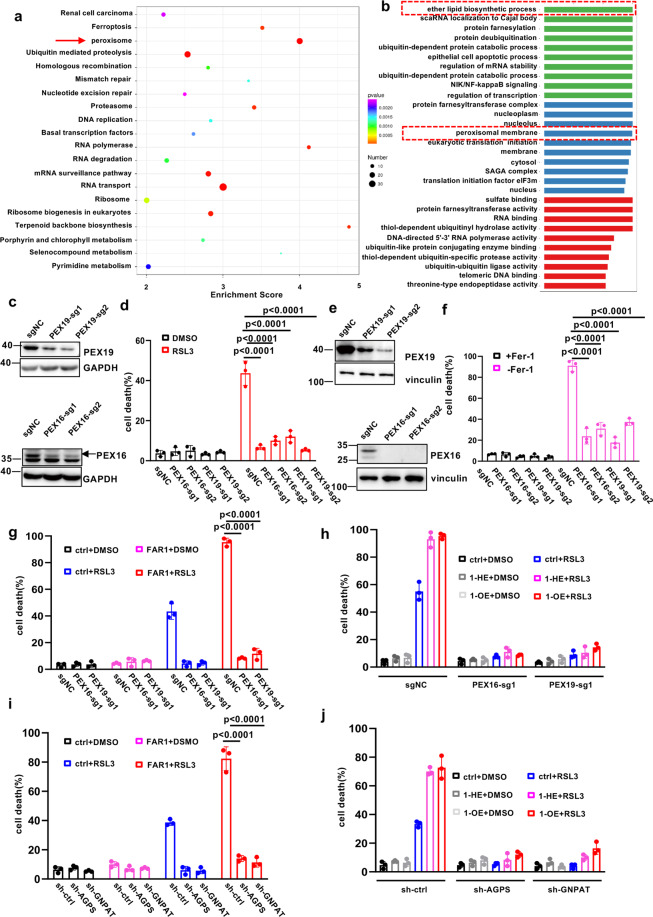
Fig. 3. FAR1-mediated ferroptosis is dependent on peroxisome-driven ether phospholipid synthesis.
a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the identified differentially expressed genes between DMSO and ML210 treated 786-O cells which are transfected with a genome-wide CRISPR-CAS9 sgRNA lentivirus pools. The twenty of most significantly enriched pathways (P < 0.05 by Fisher’s exact test) are shown, highlighting peroxisome pathway. b The Gene Ontology (GO) analysis showing that peroxisome membrane proteins and ether phospholipid biosynthetic process are involved in the process of ferroptosis. c Western blot analysis of HT1080 cells expressing sg-ctrl, sg-PEX16 or sg-PEX19. The experiments were repeated twice, independently, with similar results. d Cell death measurement of HT1080 cells expressing sg-ctrl sg-PEX16 or sg-PEX19 treated with RSL3 (500 nM) for 12 h. e Western blot analysis of HT1080 GPX4−/− cells expressing sg-ctrl, sg-PEX16 or sg-PEX19. The experiments were repeated twice, independently, with similar results. f Cell death measurements of HT1080 GPX4−/− cells expressing sg-ctrl, sg-PEX16, or sg-PEX19 upon Fer-1 withdrawal. g Cell death measurement of HT1080 cells expressing sg-ctrl sg-PEX16 or sg-PEX19 transfected with FAR1 treated with RSL3 (200 nM) for 12 h. h Cell death measurement of HT1080 cells expressing sg-ctrl sg-PEX16 or sg-PEX19 pre-incubated with 1-HE (20 μM) or 1-OE (20 μM) treated with RSL3 (500 nM) for 12 h. i Cell death measurement of HT1080 cells expressing sh-ctrl sh-GNPAT or sh-AGPS transfected with FAR1 treated with RSL3 (500 nM) for 12 h. j Cell death measurement of HT1080 cells expressing sh-ctrl sh-GNPAT or sh-AGPS pre-incubated with 1-HE (20 μM) or 1-Oct (20 μM) treated with RSL3 (200 nM) for 12 h. Data and error bars are mean ± s.d., n = 3 (d, f–i) independent repeats. All P values were calculated using two-tailed unpaired Student’s t-test.