Figure 1.
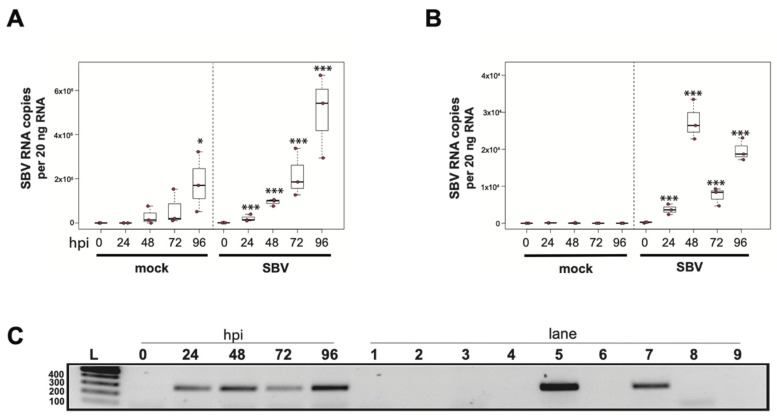
Sacbrood virus (SBV) naturally infects hemocytes and replicates in primary honey bee hemocyte cell cultures. SBV abundance was assessed in mock- or SBV-treated larval hemocytes by qPCR over a time course (i.e., 0 hpi, 24 hpi, 48 hpi, 72 hpi, and 96 hpi). (A) Cells isolated from asymptomatic larvae and treated with PBS (pH 7.4) had a 501× increase in mean SBV abundance by 96 hpi relative to 0 hpi (p = 0.042), indicating that the hemocytes in this experimental replicate had a pre-existing SBV infection and that hemocytes are a natural site of infection for SBV. The addition of filtered larval lysate containing 2.5 × 106 SBV RNA copies resulted in a 13× increase by 24 hpi relative to 0 hpi (p < 0.001) and a peak fold increase of 355× by 96 hpi (p < 0.0001). (B) In a second replicate of the experiment, mock-infected cells remained uninfected over the course of the experiment. The addition of filtered larval lysate containing 2.5 × 106 SBV RNA copies resulted in a peak fold increase of 182× (p < 0.0001) at 48 hpi, with a decrease at 72 hpi (49× relative to 0 hpi, p < 0.0001) and a subsequent rise again at 96 hpi (132× relative to 0 hpi, p < 0.0001). Raw data are included in Supplemental Table S2. All differences in means relative to 0 hpi were assessed by a Dunnett’s test. Significance levels: * p < 0.05; *** p < 0.0005. (C) To confirm that SBV was productive in infecting larval hemocytes, the presence of the negative strand (a replicative intermediate) was assessed by negative-strand-specific reverse transcription (RT) followed by PCR. Negative strand was detected at all time points after 0 hpi. Additional control reactions were performed using pooled RNA from time points 24–96 hpi (Lanes labeled 1–9). Specifically, RNA isolated from SBV containing cell lysate was reverse transcribed with the primer listed below, treated with Exonuclease I to remove excess primer, and amplified using the PCR primers listed for each lane. (L) Molecular weight ladder. (0, 24, 48, 96 hpi)—RT with random hexamers, PCR with SBV-221-240-For and SBV-478-497-Rev. (1) Negative control: No RT in the presence of (SBV-TAGS-F), PCR with TAGS and SBV-478-497-Rev. (2) Negative control: RT in the presence of (SBV-TAGS-F), PCR with only the SBV-478-497-Rev primer. (3) Negative control: RT with random hexamer primer, PCR with TAGS and SBV-478-497-Rev. (4) Negative control: No RT enzyme in the presence of random hexamers, PCR with SBV-221-240-For and SBV-478-497-Rev. (5) Positive control: RT with random hexamers, PCR with SBV-221-240-For and SBV-478-497-Rev. (6) Negative control: RT with random hexamers, PCR with only SBV-478-497-Rev primer. (7) Self-priming: RT without a primer, PCR with SBV-221-240-For and SBV-478-497-Rev. (8) Negative control: No template, no RT, PCR with TAGS and SBV-478-497-Rev. (9) Negative control: No template, no RT, PCR with SBV-221-240-For and SBV-478-497-Rev. Additional details provided in Supplemental Table S25. When reverse transcription was performed without primer, SBV cDNA was detectable (lane 7), indicating that the secondary structure in the RNA genome may serve as a primer for cDNA synthesis.