Fig. 1.
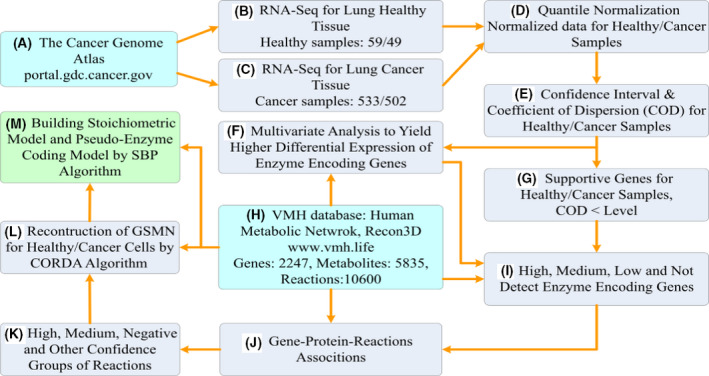
Roadmap of the reconstruction of genome‐scale metabolic models. Roadmap of the reconstruction of genome‐scale metabolic models for LUAD and LUSC and their corresponding healthy tissues. (A) Download RNA‐Seq data of LUAD and LUSC from the TCGA database. (B–G) Statistical analysis of download RNA‐Seq data to generate input information for the CORDA algorithm. (H) The general human GSMN (Recon 3D) was downloaded from VHM database (https://www.vmh.life) and used as a base model. (I) Classify enzyme‐encoding genes into four classes. (J) Compute gene–protein–reaction associations using the enzyme‐encoding genes and Recon 3D general model. (K) Identify reactions having various confidence indices. (L) Reconstruct a tissue‐specific metabolic model using the CORDA algorithm and Recon 3D general model. (M) Build the stoichiometric and GPR models in GAMS format for simulation.
