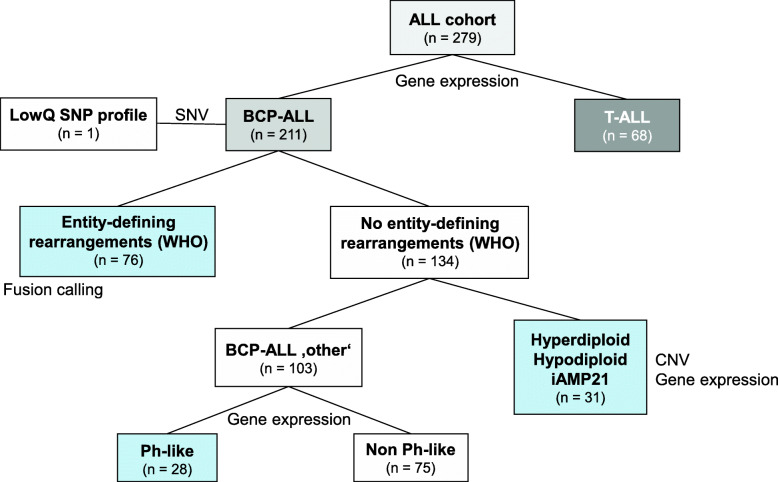
Fig. 1.
Classification tree. Design of the stepwise classification approach and distribution of the patients. First, DNA and RNA-based genotyping (SNV) was used to ensure correct WTS-WGS pairing, excluding one sample with low concordance. Gene expression analysis was used to distinguish between BCP-ALL or the T-ALL group. Subclassification of BCP-ALL samples was done progressively, assessing first the presence of entity-defining rearrangements (WHO) by the means of fusion detection. Next, copy number variations were inferred from WTS data to identify relevant ploidy groups. Lastly, gene expression profiling was used to identify BCR-ABL1-like signatures. ALL: Acute lymphoblastic leukemia; BCP-ALL: B-cell precursor acute lymphoblastic leukemia; CNV: copy number variation; iAMP21: intrachromosomal amplification of chromosome 21; LowQ: low quality; Ph: BCR-ABL1; SNP: single nucleotide polymorphism; T-ALL: T-cell acute lymphoblastic leukemia; WHO: world health organization