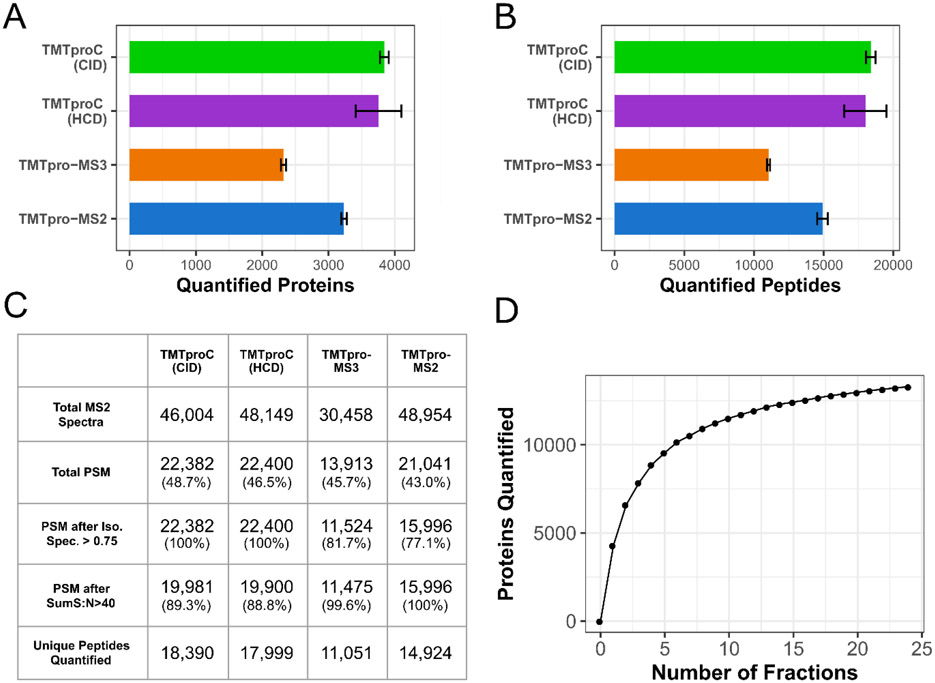
Figure 4. Evaluating TMTproC sensitivity for the analysis of a sample proteome.
A, B) Number of quantified HeLa proteins (A) and peptides (B) in replicates of a 120 min unfractionated analysis of a TMTpro0-tagged HeLa lysate sample. Both TMTproC measurements were analyzed five times, while TMTpro-MS2 and MS3 quantification were each done four times. Error bars show a single standard deviation from the mean. TMTproC using CID fragmentation is more sensitive than either TMTpro-MS3 or TMTpro-MS2 in the number of quantified peptides and proteins (Student’s t-test p-value<10−3 for all four comparisons). The sensitivity of TMTproC using HCD fragmentation also outperforms TMTpro-MS3 and TMTpro-MS2, and was not significantly different from CID fragmentation (Student’s t-test p-value=0.60 at the peptide level and p-value=0.61 at the protein level). C) Average peptide spectral matches (PSM), quantifications and effects of filter criteria for the analyses in A) and B). Percentages in parentheses in each cell represent the proportion of PSM that passed that filter. With TMTproC, isolation specificity filters do not have to be applied due to higher interference resistance, but stringent sum S:N thresholds for the ions used for quantification lead to some peptide removal. Still, after filtering, TMTproC outperforms the reporter ion methods in both number of quantified peptides and overall quantification rate. D) Number of human and yeast proteins quantified from a mixed sample shot with TMTproC (S:N>40, 1% Protein FDR, max ppm deviation<10) as a function of the number of fractions used. Only a handful of fractions were necessary to quantify more than 10,000 proteins, and the full 24 fractions led to 13,960 quantified proteins.