Extended Data Fig. 8|. The H3K27me3-binding-defective mutation of BAHCC1BAH slowed growth of acute leukemia in vitro and in vivo.
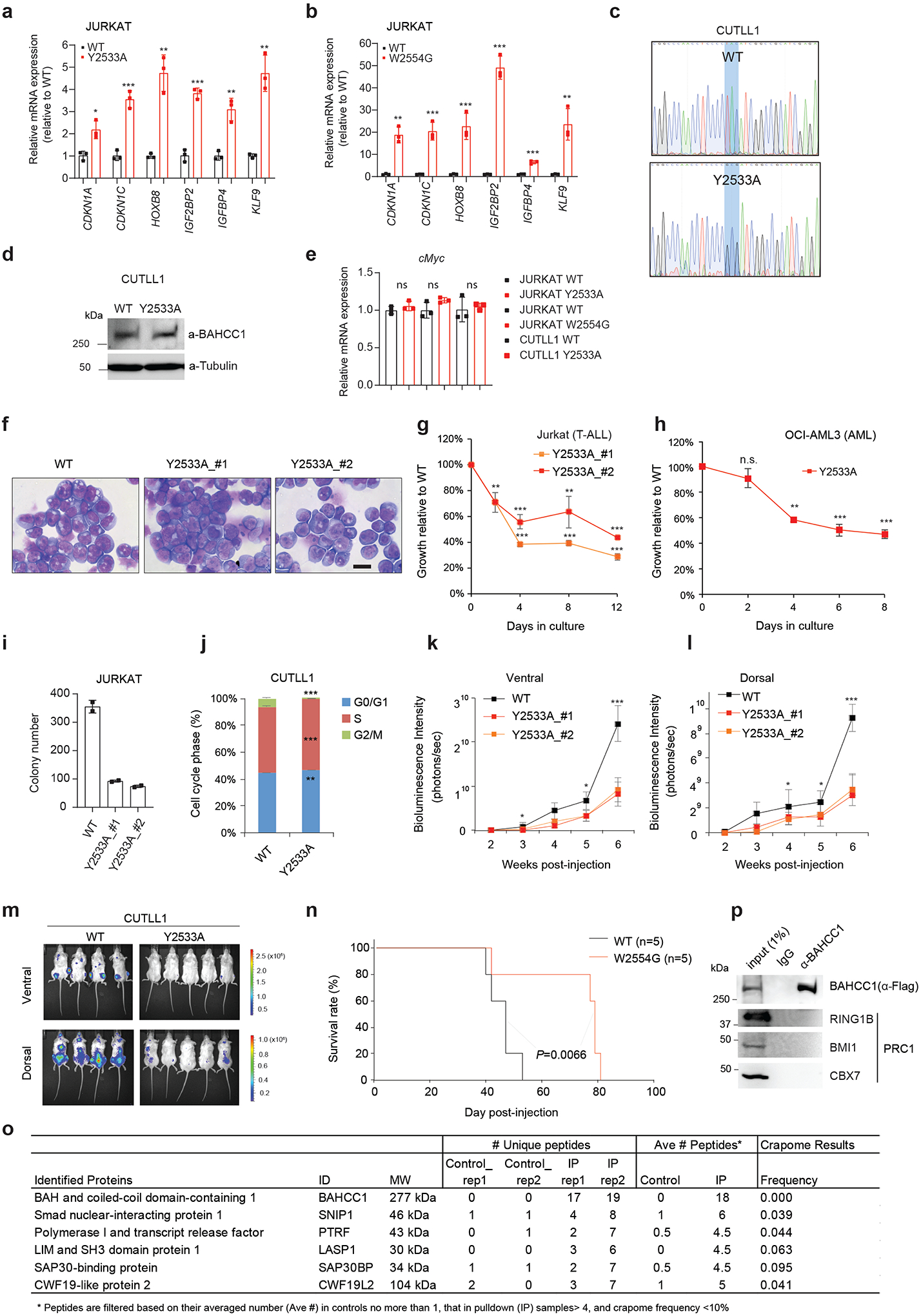
a-b, RT-qPCR of H3K27me3-marked genes in JURKAT cells harboring Y2533A (a) or W2554G (b) homozygous mutation of BAHCC1BAH, relative to WT (n=3 independent experiments). Data are plotted as mean ± SD after normalization to GAPDH and to WT. *P < 0.05; **P < 0.01; ***P < 0.001.
c-d, Sanger sequencing (c) and Flag immunoblotting (d) using CUTLL1 cells with either WT or Y2533A homozygous mutation of endogenous 3×Flag-BAHCC1.
e, RT-qPCR of cMyc in JURKAT and CUTLL1 cells with the indicated BAHCC1BAH homozygous mutation relative to WT (n=3 independent experiments). Data are plotted as mean ± SD after normalization to GAPDH and to WT. n.s., not significant.
f, Wright-Giemsa staining of JURKAT cells with WT or the Y2533A homozygous mutation of BAHCC1BAH (n=2 independent lines). Scale bar, 10 μm.
g-h, Proliferation of JURKAT (g) or OCI-AML3 cells (h) carrying homozygous mutation of BAHCC1BAH relative to WT (n=3 biologically independent experiments). Data are presented as mean ± SD.
i, Quantification of colonies formed by JURKAT cells carrying WT or Y2533A homozygous mutation of BAHCC1 (n=2 biologically independent experiments). Data are presented as mean values ± SD.
j, Quantification of cell cycle progression of CUTLL1 cells carrying WT or Y2533A homozygous mutation of BAHCC1 (n=3 biologically independent experiments). Data are presented as mean ± SD. **P < 0.01; ***P < 0.001.
k-l, Averaged bioluminescence signals collected from ventral (k) or dorsal view (l) of NSG mice (n=5) xenografted with luciferase-labeled JURKAT cells that carry WT or the Y2533A homozygous mutation (two independent lines) of BAHCC1BAH. Data are presented as mean ± SD. *P < 0.05; ***P < 0.001.
m, Bioluminescence images (two weeks post-xenograft) of NSG mice xenografted with luciferase-labeled CUTLL1 cells that carry WT or the Y2533A homozygous mutation of BAHCC1BAH.
n, Kaplan-Meier curve showing event-free survival of NSG mice xenografted with luciferase-labeled JURKAT cells that carry WT or the W2554G homozygous mutation of BAHCC1BAH. n, cohort size.
o, Summary for top hits of the BAHCC1-interacting proteins identified by Flag pulldown and mass spectrometry analyses with HeLa cells carrying BAHCC1–3×Flag KI alleles. Parental HeLa cells were used as control for Flag pulldown (n=2 biologically independent experiments). Rep, replicate.
p, CoIP for endogenous Flag-BAHCC1 and PRC1 in JURKAT cells.
