Extended Data Fig. 9|. BAHCC1 interacts with corepressors SAP30BP and HDAC1, maintaining a hypoacetylated state at target genes.
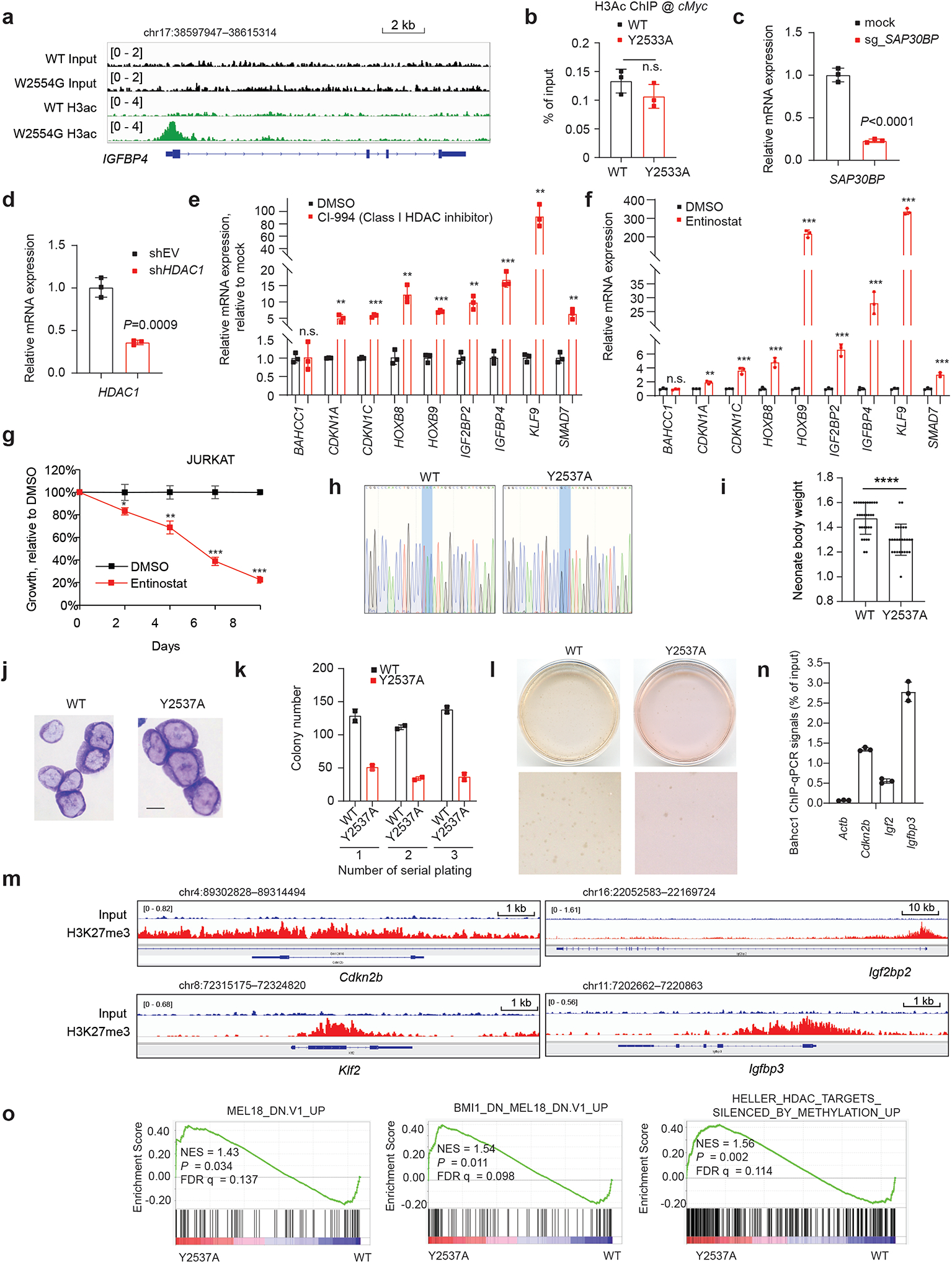
a, ChIP-seq profile of histone acetylation at the BAHCC1 target gene IGFBP4 in JURKAT cells carrying WT or the W2554G homozygous mutation of BAHCC1BAH.
b, ChIP-qPCR of histone acetylation at the cMyc promoter in JURKAT cells that carry WT or the Y2533A homozygous mutation of BAHCC1 (n=3 biologically independent samples). Data are presented as mean ± SD.
c-d, RT-qPCR of SAP30BP or HDAC1 after the dCAS9-KRAB/sgRNA-mediated repression of SAP30BP (c) or transduction of HDAC1-targeting shRNAs (d), relative to mock, in JURKAT cells (n=3 biologically independent samples). Data are presented as mean ± SD.
e-f, RT-qPCR of BAHCC1-targeted genes post-treatment of JURKAT cells with 5 μM of CI-994 (e) or 0.25 μM of Entinostat (f) for 2 days (n=3 biologically independent samples). Data are presented as mean values ± SD. **P < 0.01; ***P < 0.001; n.s., not significant.
g, JURKAT cell proliferation post-treatment with 0.25 μM of Entinostat relative to mock (n=3 biologically independent experiments). Data are presented as mean ± SD. * P < 0.05; **P < 0.01; ***P < 0.001.
h, Sanger sequencing to confirm the mouse genotype, either WT (left) or Y2537A-mutated (right) Bahcc1 allele.
i, Averaged body weight of mouse pups with either WT (left, n=31) or the Y2537A homozygous alleles (right, n=26) of Bahcc1, at day 1 post-birth (P1). Data are presented as mean ± SD. ****P < 0.0001.
j, Wright-Giemsa staining of E2A-PBX1-transformed murine leukemia cells, established with HSPCs from mice carrying either WT or the Y2537A homozygous mutation of BAHCC1BAH. Scale bar, 10 μM.
k-l, CFU quantification (k) and representative plate images (l) using the indicated E2A-PBX1-transformed murine leukemia cells (duplicate per group; data presented as mean ± SD).
m, ChIP-qPCR detects Bahcc1 binding to TSS of H3K27me3-marked genes relative to a negative locus, beta-actin (left), in the WT E2A-PBX1-transformed murine leukemia cells (n=3 independent experiments; mean ± SD after normalization to input).
n, H3K27me3 ChIP-seq profile of the indicated gene in the WT E2A-PBX1-transformed leukemia cells.
o, GSEA reveals that, relative to WT, the Y2537A mutation of Bahcc1BAH is positively correlated to derepression of Polycomb- or HDAC-related gene signature.
