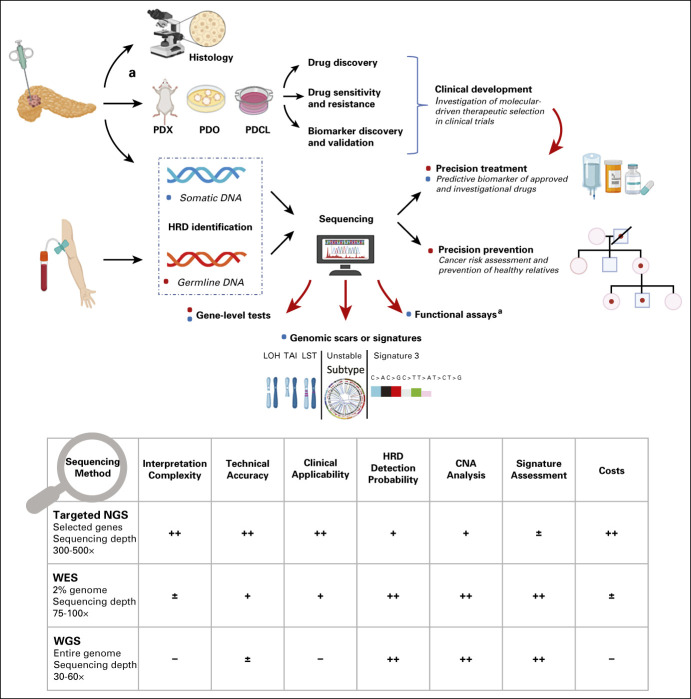
FIG 2.
Overview of HRD identification and clinical implications. Although WGS represents the most comprehensive method for HRD identification as it delivers integrated analyses of all genomic events, many barriers limit its utilization in the clinic, feasibility of accessing fresh biopsy material of sufficient size, cost, and analytic complexity. WES is a more accessible strategy and is often proposed as the second choice. However, it seems not to be the optimal method for cancer profiling as many driver events occur outside the coding exome may be missed, on one hand, and the majority of included genes are not cancer genes, on the other. Despite some technical limitations, targeted-capture sequencing delivering comprehensive genomic information, including individual gene mutations, signatures, and structural variation patterns, may represent a reasonable option for real-world applicability (practical and financial advantages compared with WES and WGS). Rating level of sequencing technologies: ++, optimal; +, good; ±, low; –, poor. aFunctional assays for real-time HRD status require in vivo or in vitro experiments. CNA, copy-number alterations; HRD, homologous recombination deficiency; LOH, loss of heterozygosity; LST, large-scale transitions; NGS, next-generation sequencing; PDCL, patient-derived cell lines; PDO, patient-derived organoids; PDX, patient-derived xenograft; TAI, telomeric allelic imbalance; WES, whole-exome sequencing; WGS, whole-genome sequencing.