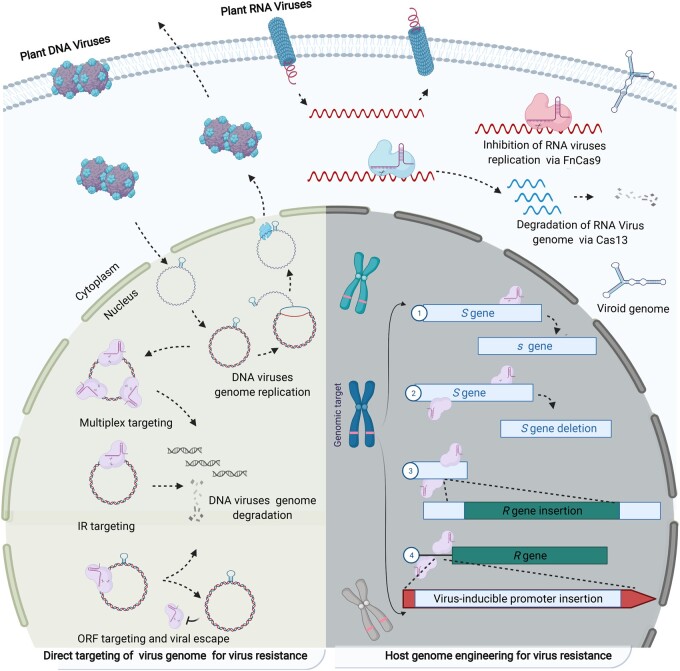
Figure 1.
Schematic of CRISPR/Cas-mediated resistance to plant viruses. Left, CRISPR/Cas-based direct targeting and interference with the genome of phytopathogenic viruses. Multisite targeting or targeting the conserved inverted repeat (IR) region provides resistance by cutting the genome of DNA viruses into multiple pieces inside the plant nucleus. Targeting plant virus genomes at the open reading frame (ORF) sequence can lead to virus escape via evolution of new ORF sequence variants that can complete the virus life cycle but are not targeted by the existing CRISPR/Cas system. Similarly, inhibition of RNA virus genome replication or degradation of the RNA genome was achieved via FnCas9 and Cas13 variants in the cytoplasm. CRISPR/Cas systems can also be designed to target the RNA genomes of viroids. Right, genetic engineering of susceptibility (S) and resistance (R) genes in the host genome can provide durable resistance to multiple viruses. CRISPR/Cas editing of S genes mimics natural resistance (point repair). Removal of S genes and insertion of R genes into neutral, high-expression loci or under the control of strong promoters, including virus-inducible promoters, via homology-directed repair can result in over-expression of R genes for virus resistance.