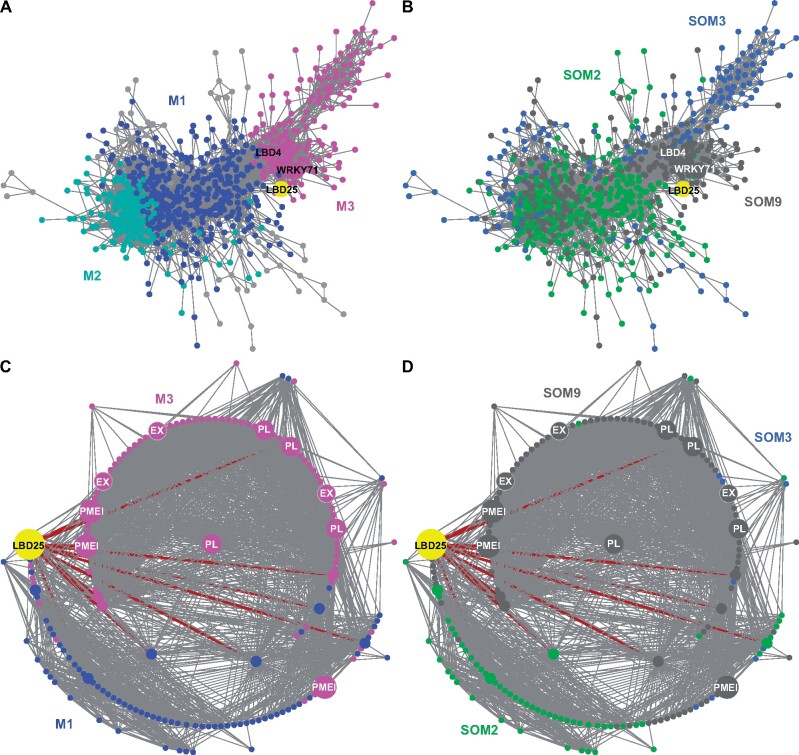
Figure 3.
GCNs of SOM2, 3, 9 genes based on C. campestris tissue type RNA-seq data. A, GCN of genes that are in SOM2, SOM3, and SOM9 with colors based on network modules. This SOM2+SOM3+SOM9 GCN is composed of three major modules. Blue indicates genes in Module 1, which has the biological process GO enrichment “morphogenesis of a branching structure, plant organ formation, strigolactone responses, and biosynthetic processes.” Cyan indicates genes in Module 2, which has the biological process GO enrichment “response to karrikin, hormone-mediated signaling pathway and defense response.” Magenta indicates genes in Module 3, which has the biological process GO enrichment “plant-type cell wall loosening.” Light grey indicates genes that are not included in Modules 1, 2, or 3. B, GCN of genes that are in SOM2, SOM3, and SOM9, with colors based on SOM clustering groups. Green indicates genes in SOM2. Blue indicates genes in SOM3. Grey indicates genes in SOM9, which includes the genes that are highly expressed in both prehaustoria and haustoria. SOM2 includes genes that are only highly expressed in haustoria and SOM3 includes genes that are only highly expressed in prehaustoria. C, GCN of CcLBD25 and its first and second layer neighbors with colors based on network modules as in (A). D, GCN of CcLBD25 and its first and second layer neighbors with colors based on SOM clustering groups as in (B). C and D, Dark red lines indicate the connection between CcLBD25 and its first layer neighbors. The genes that are second-layer neighbors of CcLBD25 are labeled with medium size dots. The genes that are outside the second layer neighbors of CcLBD25 are labeled with small size dots. CcLBD25 and cell wall loosening related genes are enlarged, highlighted, and labeled in the network.