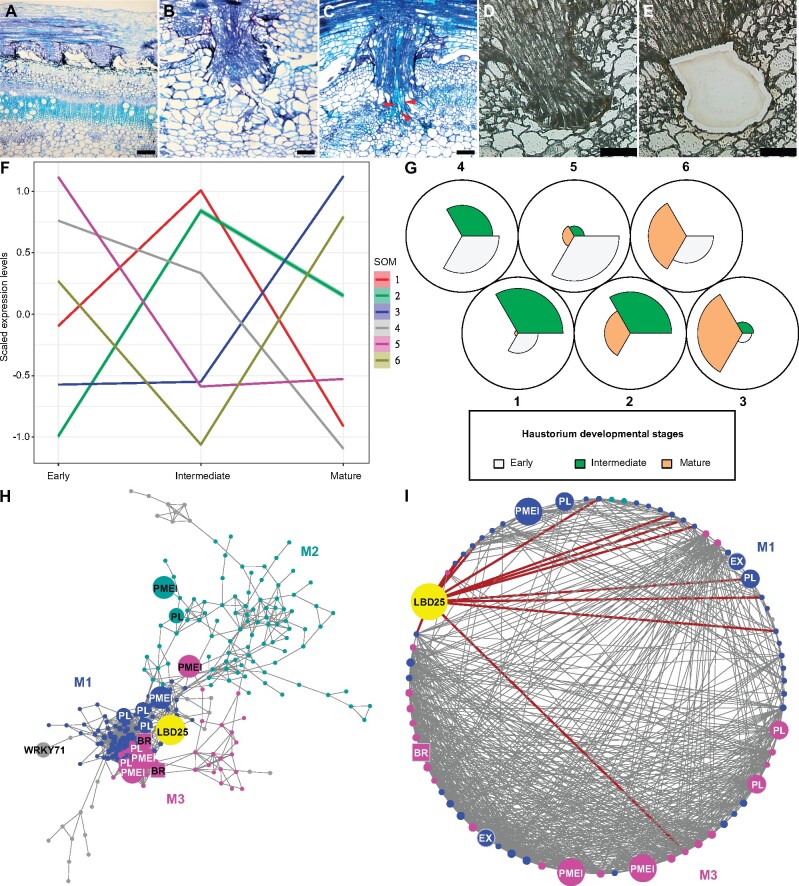
Figure 4.
SOM clustering and GCNs of gene expression in C. campestris haustoria across three developmental stages (from the LCM RNA-seq data). A–E, Sections of developmental stages for LCM RNA-seq. A–C, Three developmental stages for LCM RNA-seq. Paraffin sections stained with Toluidine Blue. A, Early stage. B, Intermediate stage. C, Mature stage. The red arrowheads indicate vascular connections between host and C. campestris. D–E, The C. campestris haustorium tissues were collected using LCM. A and C, Scale bars = 250 µm. B, D, and E, Scale bars = 100 µm. F, Scaled expression levels of each SOM group across three haustorium developmental stages. Each line is colored based on the corresponding SOM group. The highlighting around the lines indicates a 95% confidence interval. G, A code plot of SOM clustering illustrating which developmental stages are highly represented in each SOM group by the size of sectors. Each sector represents a developmental stage and is in the color indicating its corresponding developmental stage. H, GCN based on LCM RNA-seq expression profiles with genes in tissue type RNA-seq SOM9. Blue indicates genes in Module 1, which has enriched biological process GO term “plant-type cell wall loosening.” Cyan indicates genes in Module 2, which has enriched biological process GO term “respiratory burst.” Magenta indicates genes in Module 3, which has enriched biological process GO term “brassinosteroid mediated signaling pathway.” Light grey indicates genes that are not included in Modules 1, 2, or 3. CcLBD25, CcWRKY71, and pectin degradation-related genes are enlarged, highlighted, and labeled in the network. The genes involved in brassinosteroid signaling are labeled in squares. I, GCN of CcLBD25 and its first and second layer neighbors with colors based on network modules as in (H). Dark red lines indicate the connection between CcLBD25 and its first layer neighbors. CcLBD25 and cell wall loosening related genes are enlarged, highlighted, and labeled in the network. H–I, PL.