Abstract
Upon returning from deployment to Afghanistan, a substantial number of U.S. military personnel report deployment-related lung disease (DRLD) symptoms, including those consistent with an asthma-like airways disease. DRLD is thought to be caused by prolonged inhalation of toxic desert particulate matter, which can persist in the postdeployment setting such as exposure to common household allergens. The goal of this study was to define the transcriptomic responses of lung leukocytes of mice exposed to Afghanistan desert particulate matter (APM) and house dust mite (HDM). C57BL/6 mice (n = 15/group) were exposed to filtered air or aerosolized APM for 12 days, followed by intranasal PBS or HDM allergen challenges for 24 h. Bronchoalveolar lavage (BAL) cells were collected for single-cell RNA sequencing (scRNAseq), and assessment of inflammation and airway hyper-responsiveness. Unsupervised clustering of BAL cell scRNAseq data revealed a unique monocyte population induced only by both APM and allergen treatments. This population of monocytes is characterized by the expression of genes involved in allergic asthma, including Alox15. We validated Alox15 expression in monocytes via immunostaining of lung tissue. APM pre-exposure, followed by the HDM challenge, led to significantly increased total respiratory system resistance compared with filtered air controls. Using this mouse model to mimic DRLD, we demonstrated that inhalation of airborne PM during deployment may prime airways to be more responsive to allergen exposure after returning home, which may be linked to dysregulated immune responses such as induction of a unique lung monocyte population.
Keywords: particulate matter, allergic asthma, ALOX15, single-cell RNA sequencing
Nearly 3 million U.S. military personnel have been deployed to Afghanistan and Iraq since the start of post-September 11, 2001 combat operations. Most had no pre-existing lung disorders, but estimates suggest that up to14% of the postdeployment military population report symptoms consistent with a deployment-related lung disease (DRLD) (Szema et al., 2010). These new-onset respiratory symptoms include exercise-induced shortness of breath, coughing, wheezing, and/or chest tightness (Krefft et al., 2020; Szema et al., 2014, 2017). The development of chronic lung diseases such as asthma and bronchiolitis has also been reported (Zell-Baran et al., 2018).
We propose that a major contributing factor to the development of DRLD is repeated, prolonged inhalation of toxic desert particulate matter (PM), which includes both airborne and sand/dust components. Previously, PM exposures from a number of outdoor pollutant sources have been linked to asthma exacerbations and asthma-like symptoms (Orellano et al., 2017; Son et al., 2013). The composition of PM varies by region, and Afghanistan PM (APM) has been reported to be composed of sand, silt, and clay, along with other metals and toxins (Engelbrecht et al., 2009). Byproducts of war may also settle in the dust, including diesel exhaust particles, burn pit debris, and debris from improvised explosive device blasts (Szema et al., 2017; Zell-Baran et al., 2018). These complex PM mixtures are aerosolized in the frequent dust storms that plague the area (Szema et al., 2010, 2011). Air quality in deserts of Southwest Asia, including Afghanistan, often exceeds the 15 µg/m3 guidelines set by the United States Environmental Protection Agency (Engelbrecht et al., 2009; Invernizzi et al., 2004; Krefft et al., 2020; Weese and Abraham, 2009; Zell-Baran et al., 2018). A study by the Canadian Armed Forces Deployable Health Hazard Assessment Team sampled PM2.5 and PM10 over 24 h at various sites in Afghanistan and applied EPA guidelines for Air Quality Index (AQI). The researchers found that the average levels of PM2.5 measured over the study period fell into the moderate to unhealthy range of AQI (12.1–34.5 and 55.5–150.4 µg/m3, respectively) and that PM10 averages corresponded to a hazardous AQI (150.5–250.4 µg/m3) (Lalonde and Bradley, 2015; Minnesota Pollution Control Agency). We previously reported that underlying, low-grade type 2 inflammation may predispose some soldiers to develop DRLD (Berman et al., 2018). However, why some soldiers continue to report symptoms of DRLD long after their tour of duty concludes remains to be explored.
After prolonged exposure to high levels of desert PM in Afghanistan, soldiers may return home and be faced with exposure to another common health hazard—household dusts containing allergens and other potentially sensitizing material. We propose that these chronic APM exposures during deployment may prime airways to be more sensitive to allergens after returning home. In studies of other particulates and ovalbumin (OVA), a synthetic allergen, animals that were sensitized then challenged with particulates and allergen at the same time developed characteristics of asthmatic inflammation (Herbert et al., 2013). Mice exposed to particulates and allergens demonstrate enhanced neutrophilic and eosinophilic inflammation, which, together with airway hyper-responsiveness (AHR)/airway narrowing, are characteristic features of asthma (Herbert et al., 2013; Zhang et al., 2018).
In this manuscript, we explore the hypothesis that exposure to household aeroallergens in airways previously exposed to APM prolongs symptoms of DRLD after the deployment ends. To mimic the postdeployment setting, we employed a repeated APM pre-exposure model followed by sensitization and challenge with house dust mite (HDM). We used single-cell RNA sequencing (scRNAseq) to investigate the diverse cell populations in bronchoalveolar lavage (BAL) fluid of mice that were treated in a manner that mimics the postdeployment setting. scRNAseq allowed us to capture the full transcriptomic response of each cell type in BAL in this postdeployment model. Ultimately, we identified a unique population of cells that may contribute to the regulation of DRLD.
MATERIALS AND METHODS
Characterization of Afghanistan Desert PM
Topsoil from Bagram Air Force Base in the Parwan Province of Afghanistan was collected in August 2009. Characterization of the sample was performed by the United States Geological Survey (USGS) in Lakewood, Colorado and described previously (Berman et al., 2018). In our sample of APM, 89.46% of particles were less than 2.5 μm in diameter (fine), 10.37% was between 2.5 and 10 μm (coarse), and 0.17% was greater than 10 μm in diameter (large). In vivo exposures were performed using bulk topsoil material that was filtered to be of a respirable size using a dry powder generator (Wright Dust Feeder, CH Technologies, Westwood, New Jersey).
In Vivo Afghanistan PM and HDM Exposures
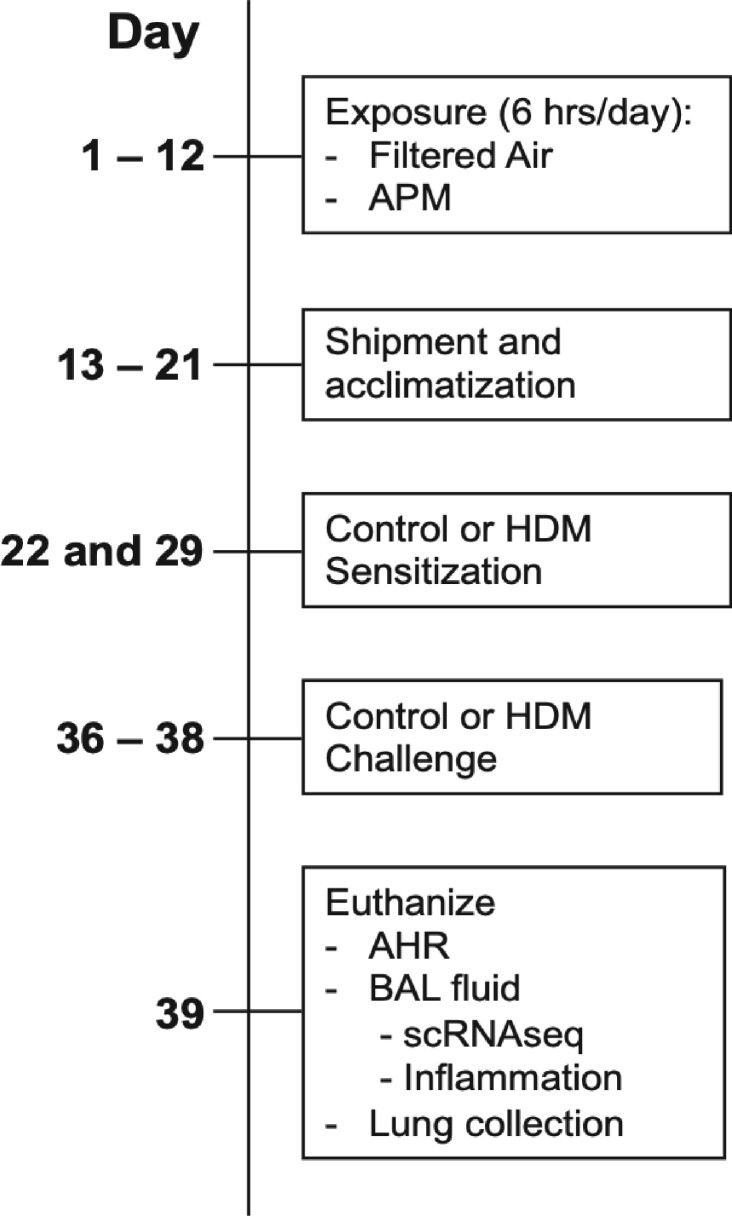
Eight-week-old C57BL/6 mice were obtained from Jackson Laboratories and housed individually under pathogen-free conditions and fed a standard chow diet. The Naval Medical Research Unit Dayton (NAMRU-D) at the Wright Patterson Air Force Base (Dayton, Ohio) first performed the whole body PM exposures, and the allergen instillations were conducted at National Jewish Health (NJH) in Denver, Colorado. This study was approved by the Institutional Animal Care and Use Committees (IACUC) at NJH and by ACURO at NAMRU-Dayton. An overview of the animal exposures is shown in Figure 1. Mice of similar age and body weight were randomized into control (filtered air) or treatment (aerosolized APM [5 mg/m3]) groups and exposed for 6 h per day for 12 consecutive days, via whole-body exposure chambers (n = 30/group) at the NAMRU-D facility as described previously (Sterner et al., 2020). After the last APM exposure, mice were shipped to NJH where they were allowed to acclimatize. Nine days after the last APM exposure, the allergen sensitizations were initiated. On days 22 and 29 of the study, mice were sensitized to either 10 μg of HDM suspended in PBS or sterile PBS (control) or via intranasal instillation (n = 15/group). On days 36, 37, and 38, mice were challenged with either PBS or HDM. All mice were exposed on each treatment day and the study represents one complete set of exposures. On day 39, mice from each group were anesthetized for analysis of airway physiology on the flexiVent (n = 8/group) or euthanized and the lungs were lavaged and cells pooled for single-cell RNAseq (n = 7/group). All 60 mice from the initial filtered air and APM exposures survived until the final study endpoint and no mice were excluded from postharvest analysis. All researchers were aware of group allocation at each stage of the experiment, except for the team performing the scRNAseq, who were blinded until the early stages of the data analysis.
Figure 1.
Overview of in vivo mouse APM and allergen exposures. C57BL/6 mice were exposed to aerosolized Afghanistan PM (APM) at a concentration of 5 mg/m3 or filtered air (FA, control) for 6 h per day on days 1–12 days. On days 22 and 29, mice were sensitized with PBS (control) or house dust mite (HDM, 10 μg/mouse) then challenged with PBS or HDM on days 36–38. Mice were euthanized on day 39.
Evaluation of Pulmonary Function
Pulmonary function was assessed using the flexiVent small animal physiology system (SCIREQ, Quebec, Canada). Animals were anesthetized for airway physiology using a mixture of ketamine (100 mg/kg), xylazine (15 mg/kg), and Acepromazine (10 mg/kg) per body weight. Once ventilation began, Pancuronium (1 mg/kg) was administered to paralyze all muscle contraction. After completion of the physiology assessment, animals were euthanized via major organ harvest. Mice were anesthetized using a cocktail of Ketamine, Xylazine, and Acepromazine, then tracheostomized and ventilated at a rate of 150 breaths/min. Full body paralysis was induced using Pancuronium with heart rate and body temperature monitored throughout the analysis. Each mouse was exposed to increasing doses of nebulized methacholine (Mch) in H2O: 0, 6.25, 25, and 50 mg/ml. Deep inflation of lungs was performed between each dose to allow airways to return to baseline. Total respiratory system resistance (Rrs) was calculated from a maximum value out of 12 measurements. After the highest dose of Mch, mice (n = 8) were immediately lavaged with 1 ml of sterile saline to collect bronchoalveolar lavage fluid (BALF). BALF was centrifuged at 6000 RPM for 5 min at 4C and cell-free supernatant was removed and frozen for later analysis of protein levels via enzyme-linked immunosorbent assay (ELISA). Cell pellets were resuspended in saline to make cytospin slides for total and differential leukocyte counts using the Diff-Quick Stain Kit (IMEB, San Marcos, California).
Single-Cell RNA Sequencing of BAL Cells
BAL cell processing for scRNAseq
Mice that were not evaluated on the flexiVent were euthanized with pentobarbital (Vortech Pharmaceuticals, Dearborn, Michigan). Fresh BAL cells were obtained by lavage of lungs with PBS, with minimal damage to the tracheas to avoid red blood cell contamination. Each mouse was lavaged 5 times and the solutions were pooled by group. BAL was kept on ice between collection and sequencing, which occurred the same day. Cells were never frozen prior to sequencing. To remove mucus in the BAL preparation, cells were resuspended in PBS and passed through 70 µm and then 40 µm pore size cell strainers. Cytospin slides were used to confirm that the cells were in a single-cell suspension prior to sequencing. The viability of cells as evaluated by trypan blue staining was between 60% and 80%.
scRNAseq library preparation
Cells were sequenced at 50 000 reads per cell within hours of harvest. scRNAseq libraries were prepared with the 10× Genomics 3ʹ-end mRNA expression kit (version 2) at the University of Colorado Genomics and Microarray Core. The processed libraries were sequenced on the Illumina NovaSeq6000 instrument paired-end 151 basepair sequencing. The scRNAseq experiment was performed with Control (filtered air + PBS), HDM (filtered air + HDM), APM (APM + PBS), and APM + HDM samples derived from 7 pooled mice for each experiment. Sequencing metrics, including depth, are shown in Table 1.
Table 1.
Sequencing Metrics of Pooled Mouse BAL Cells
| Treatment | No. of Cells Sequenced | Median UMIs | Median Genes Expressed |
|---|---|---|---|
|
Control: Filtered air + saline |
5303 | 2836 | 1283 |
|
Allergen: Filtered air + house dust mite |
1056 | 1351 | 602 |
|
APM: APM + saline |
4526 | 2441.5 | 1146 |
|
APM + allergen: APM + house dust mite |
1395 | 4621 | 1375 |
After filtered air or APM exposure, mice were challenged with PBS or HDM. One day after the last challenge, mice were euthanized and BAL was collected for scRNAseq. BAL was pooled from 7 mice/group prior to sequencing. Libraries were prepared using the 10× Genomics 3ʹ-End Microarray Expression kits and sequenced using the Illumina NovaSeq6000.UMI indicates the unique molecular identifiers or number of transcripts detected in the sample.
Bioinformatics
Fastq files were processed using cellranger (v. 2.2.0) with the mm10 genome assembly to generate unique molecular identifier (UMI) gene count matrices per sample. Seurat (v. 2.3.4) was used to aggregate all samples into a single matrix and to perform initial quality control filtering, normalization, and clustering (Butler et al., 2018). Cells were removed if there were fewer than 250 genes detected, a UMI count less than 250 or greater than 10 000, or if the proportion of UMIs mapped to mitochondrial genes was greater than 10%. Genes were excluded if they were detectable in fewer than 5 cells. Following filtering, the UMI counts were normalized to library size (total number of UMIs detected), scaled by 10 000, and log-transformed. Principal component analysis was performed on the zscores of the normalized expression values and 17 dimensions were selected for tSNE projection using a perplexity of 30. Graph-based clustering was performed using the top 17 principal components, with 30 nearest neighbors, and a resolution of 0.8 or other resolution. Cell-type clusters were named according to the gene expression signatures seen in Table 2.
Table 2.
Cell-Type Markers in Mouse BAL
| Cell Types | Markers | % of Total Cell Population |
|---|---|---|
| Macrophages | CD68, Lyz2, Marco, SerpinB2, F4/80 | 80.2 |
| Monocytes | 3.1 | |
| Neutrophils | Gr-1, Ly6G, S100a8, S100a9 | 2.1 |
| Eosinophils | Eosinophil-associated ribonuclease (EAR), eosinophil-derived neurotoxin (ECP2), CD193, Siglec-F | |
| T cells | CD3 | 7.0 |
| Natural killer cells | CD49b, CD3(–) | |
| B cells | CD19, Ly6d | 5.8 |
| Dendritic cells | CD11c | 0.77 |
| Airway epithelial cells | 1110017D15Rik, Foxj1, Scgb1a1, Scgb3a2, keratin 7, Aldh1a2 | 1.0 |
| Alveolar epithelial cells (type II) | Sftpc, Sftpa1, and Sftpb. | |
| Alveolar epithelial cells (type 1) | Pdpn, Ager, and Clic5 |
Clusters were classified by cell type using specific gene expression signatures according to Ref # (Han et al., 2018).
Following clustering and tSNE projection, UMI matrices for each sample were normalized as described above and the resulting gene expression values were used for all downstream analyses. Genes differentially expressed in each cluster compared with other clusters in each tested comparison were determined using FindAllMarkers() in Seurat, where the () indicates the dataset to be analyzed. Genes were included if they were detectable in a minimum of 10% of cells in the cluster of interest, there was at least a 0.25-fold change (natural log scale) difference in expression between the cluster of interest and the other clusters, and the p-value was < .10. Of note, because of the relatively low efficiency of current scRNAseq library preparation methods, many mRNAs, particularly those of low-moderate expression levels, are not detected in some cells in which they are present (Kharchenko et al., 2014; Zheng et al., 2017). Heatmaps and tSNE plots were generated in Seurat using the DoHeatmap() and FeaturePlot() commands. Single-cell RNASeq sequencing reads and processed data were deposited to GEO (accession number: GSE155391).
Ingenuity pathway analysis
Differential gene expression data from each cluster were uploaded to ingenuity pathway analysis (IPA, Version 01-14, Qiagen Bioinformatics). Using the MyPathway explorer function, a signaling pathway reflecting up- and downregulation of genes within the unique monocyte cluster was generated with asthma and allergic inflammation set as the pathway endpoint. A disease tree map was generated using the differential gene expression signatures of the entire cluster.
Alox15 Staining of Mouse Lung Tissue
After the mice were lavaged for scRNAseq, the lungs were harvested and fixed in 10% formalin until paraffin embedding. For colorimetric immunostaining, lungs were cut into 5-µm thick sections and dewaxed. After antigen retrieval via heating in sodium citrate buffer, blocking, and quenching of endogenous peroxidase activity, slices were incubated overnight in Alox15 antibody (rabbit anti-mouse Alox15) at a 1:200 dilution (Abcam, Boston, Massachusetts), followed by incubation with a biotinylated goat anti-rabbit IgG and avidin-biotin-peroxidase complex (Vector Lab., Burlingame, California). After rinsing the slides in TBS, 0.03% aminoethylcarbazole (AEC) in 0.03% hydrogen peroxide was used as a substrate to develop a peroxide-dependent red color reaction. Slides were counterstained with Mayer's hematoxylin and covered with Crystalmount (Biomeda Corp., Foster City, California). Slides were visualized at 200×.
Immunofluorescent Staining of Mouse Lung Tissue
We performed immunofluorescent staining of lung tissue to validate the presence of Alox15 positive monocytes that were identified using scRNAseq. Lungs were harvested after lavage for scRNAseq and fixed in 10% formalin until paraffin embedding. Lungs were cut into 5-µm sections and dewaxed using xylezene and ethanol washes. Antigen retrieval was performed using citrate buffer and heating, then slides were blocked. Slides were incubated overnight in Alox15 antibody (from rabbit) at a 1:200 dilution (ab244205, Abcam, Boston, Massachusetts) and F4/80 conjugated to APC (BM8, Invitrogen, Carlsbad, California) at a dilution of 1:50. Slides were incubated with an antirabbit secondary antibody conjugated to AF555 at a 1:100 dilution (Invitrogen, Carlsbad, California). Slides were incubated with DAPI (2 μg/ml) then mounted with fluorescent mounting media (Agilent, Santa Clara, California) and visualized using a Zeiss LSM700 confocal microscope at 40× and 63× and analyzed using Zen Black software.
Enzyme-Linked Immunosorbent Assay
Levels of Eotaxin-2 and KC protein were measured in cell-free BAL fluid collected from mice after airway physiology assessment using the ELISA Duoset Kits (R&D Systems, Minneapolis, Minnesota).
Statistical Analysis
Genes differentially expressed between clusters were determined using the Wilcoxon rank-sum test using expression values in each cluster compared with all other clusters. Differential gene expression is reported using adjusted p-values with Bonferroni correction for multiple hypothesis testing. The lower limit of detection for the p-values was 2.225074 × 10−308. The overlap p-values were calculated using Fisher’s exact test. For scRNAseq differential gene expression experiments, p ≤ .10 was considered significant (Vieth et al., 2019). A one-way ANOVA with a Tukey’s post-test was used to compare differences in means between groups when more than 2 groups are present. A p-value of less than .05 was considered significant. Figures are presented with a single bar to indicate median or error bars indicating means ± SEM.
RESULTS
Unsupervised Clustering Identifies Cell Types Based on Gene Expression
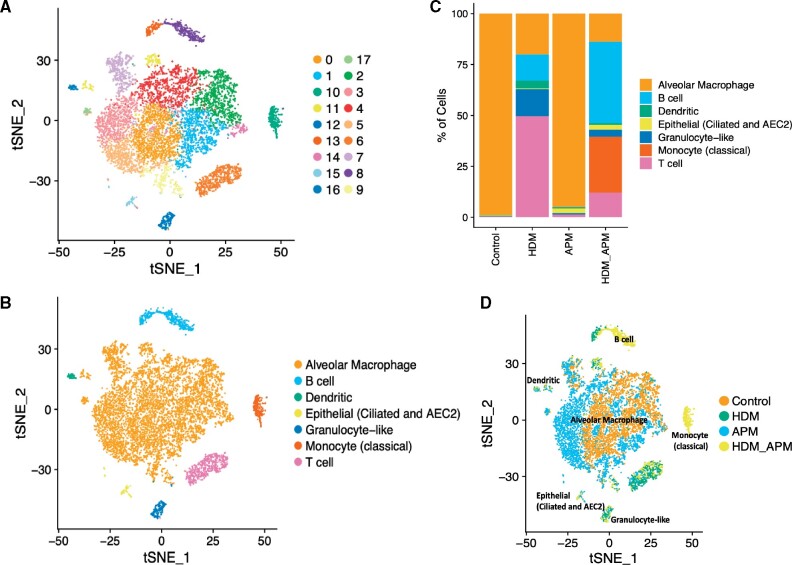
We generated scRNAseq data on 12 280 BAL cells across our 4 treatment groups—Control, HDM, APM, and APM + HDM (Table 1). Among these cells, we identified 17 transcriptionally distinct cell populations through shared nearest neighbor clustering analysis and visualized these cells by 2-dimensional embedding using tSNE (Figure 2A). Enriched gene markers for the cell clusters were identified and intersected with known cell-type markers to assign clusters a cell-type identity, as listed in Table 2 (Figure 2B) (Han et al., 2018). Combining clusters with similar cell-type markers resulted in the identification of 7 distinct cell types (Figure 2B), with alveolar macrophages as the predominant cell type in BAL cell populations, as expected. Other cell types included monocytes, T cells, and B cells. A small number of epithelial cells were found, likely due to epithelial sloughing during the BAL collection procedure. Although granulocytes, including neutrophils and eosinophils, are a common cell constituent of the BAL, these cell types were not observed due to technical limitations associated with the 10× Genomics platform and were not captured. The small observed population may be due to the presence of a common cell marker, but the majority of neutrophils and eosinophils were not captured.
Figure 2.
scRNAseq revealed unique cell populations induced by different treatments. tSNE plots of mouse BAL cells were constructed in Seurat. Unsupervised clustering of mouse BAL cells revealed 17 distinct clusters (A). Clusters were named according to cell marker genes (B). Proportions of different cell types vary across treatments, revealing a unique cluster of monocytes is induced by only one treatment—HDM and APM (C and D).
Cell Clusters and Gene Expression Differ Based on Treatment Conditions
Next, we evaluated the proportions of each type of cell identified in Figure 2B to investigate if APM + HDM treatments induce the recruitment of specific cell types into the airways. We observed that the proportions of each cell type varied depending on treatment condition, as well as the percentage of cells of that subtype per treatment condition (Figure 2C). The greatest numbers of cells in the pooled BAL sample were alveolar macrophages, divided almost evenly between control and APM conditions. The alveolar macrophage cluster was predominantly composed of cells from nonallergen-treated mice, with a very low percentage of HDM-exposed cells. Interestingly, the classical monocyte cluster of cells was induced only by APM + HDM treatment.
Clustering of Cells by Treatment Revealed Distinct BAL Cell Populations
To further examine the effect of each treatment on BAL cellular distribution, we overlaid treatment identity on the tSNE plot (Figure 2D). By labeling the clusters by cell type and treatment condition, we found a unique monocyte cluster induced only by the combination of APM followed by HDM. This population of monocytes is characterized by a gene expression signature of 606 genes, many of which belong to pathways involved in the worsening of asthma-like airways diseases. These include Alox15 (Arachidonate 15-Lipoxygenase, 15-LOX), which is involved in airway obstruction; Itgam (Integrin Subunit Alpha M), which mediates inflammation via regulating leukocyte adhesion/migration; and Ccl24 (Eotaxin-2), which induces recruitment of eosinophils. Additional genes with the signatures of M2 macrophages that are associated with cell proliferation and repair, including Serpinb2, Arginase 1, and CCL9, were also enriched in this cluster. A heatmap of the gene expression within the monocyte cluster that was induced only by APM + HDM reveals the most enriched genes within this cluster (Figure 3A). Of particular interest is Alox15, which is found only within this population of cells. Using a starting point of Alox15 in an asthmatic context, we built a signaling pathway using IPA software. This pathway demonstrated enrichment of downstream targets of Alox15 that are associated with allergic asthma including FN1 (Fibronectin-1), ITGAM, and C3 (Complement 3) (Figure 3B). Fold changes and gene expression by cell and cluster are seen in Table 3. We were next able to visualize that the expression of Alox15 is highly expressed in the monocyte cluster when looking at the overall cluster of cells (Figure 3C).
Figure 3.
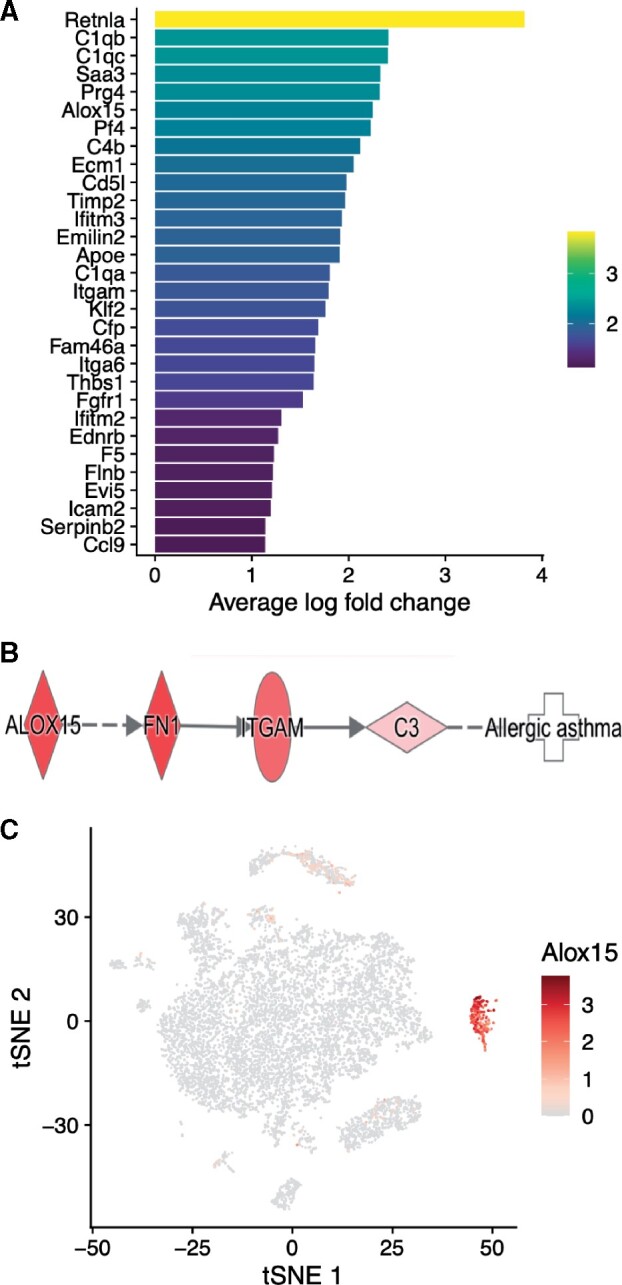
Combination of APM and HDM treatment induces expression of allergic asthma-associated genes in the monocyte cluster. Heatmap of top genes expressed in the monocyte cluster, induced by APM + HDM treatment (A). Yellow indicates upregulation and purple indicates downregulation, presented as average log fold change. In (B), a signaling pathway of genes involved in allergic asthma shows how Alox15 and other genes in the monocyte cluster contribute to allergic asthma. Alox15 is expression is concentrated in the monocyte cluster (C). A tSNE plot colored by Alox15 expression demonstrates that Alox15 gene expression is concentrated highly in cells of the monocyte cluster, which is induced only by combination APM + HDM treatment. Alox15 expressing cells are shown on the spectrum of yellow to red, gray indicates no expression.
Table 3.
Fold Changes for Genes in Alox15 Signaling Pathway
| ID | Fold Change | Cells Expressing Gene in Cluster (%) | Cells Expressing Gene in All Clusters (%) |
|---|---|---|---|
| Alox15 | 2.24905554 | 0.968 | 0.013 |
| Fn1 | 3.11389901 | 1 | 0.314 |
| Itgam | 1.79166236 | 0.981 | 0.025 |
| C3 | 0.54306139 | 0.614 | 0.148 |
After scRNAseq, a unique monocyte cluster was revealed. IPA was used to generate a signaling pathway associated with allergic asthma induced by Alox15 with genes expressed in the monocyte cluster.
Alox15 Expressing Monocytes in Lung Tissue
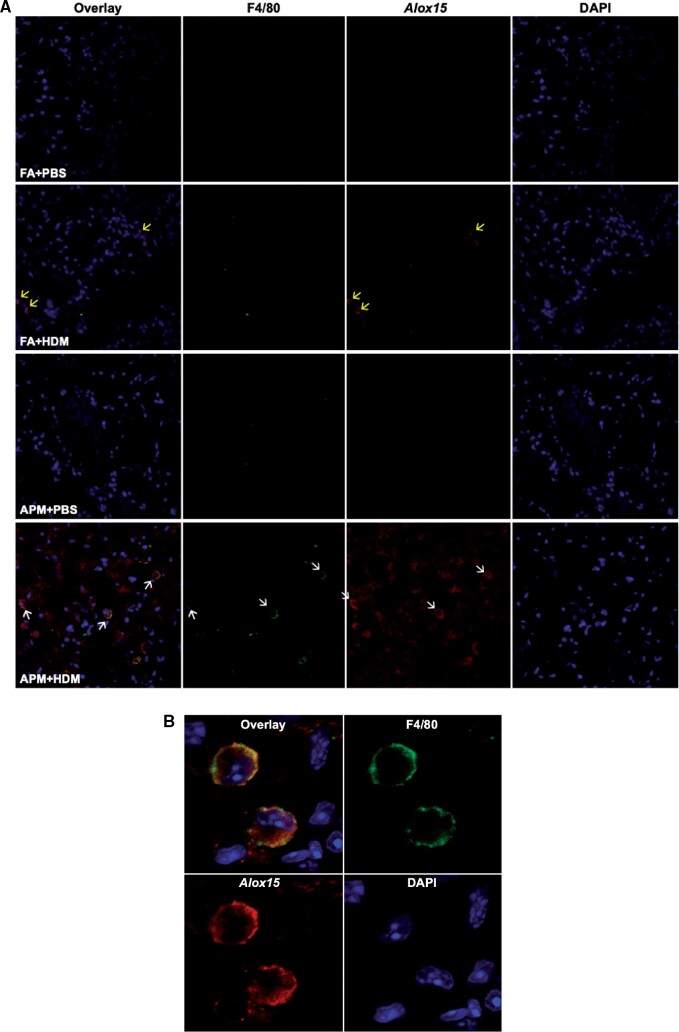
After identifying Alox15 as a gene of interest, our next objective was to validate its expression by specific cell types in the tissue. To demonstrate first that Alox15 was expressed in the lung and second, that it was expressed by monocytes, we performed dual immunofluorescent staining of lung tissue harvested after the lavages for scRNAseq. We used antibodies against Alox15 and F4/80, a monocyte/macrophage marker to further validate the scRNAseq data. Immunofluorescence imaging demonstrated that cells positive for both Alox15 (shown in red) and F4/80 (pseudocolored in green) were present only in lung tissues of APM + HDM treated mice, primarily in the alveolar space (indicated by white arrows) (Figure 4A). A close-up image of 2 monocytes taken at a higher magnification shows the costaining of both Alox15 and F4/80 (Figure 4B). In filtered air + HDM treated mice, allergen induced Alox15 expression in nonmonocyte cells (yellow arrows).
Figure 4.
Presence of Alox15 positive monocytes in lung tissue from APM + HDM exposed mice. Lung tissue was harvest after BAL collection and stained with F4/80, a monocyte/macrophage marker (conjugated to APC 647, pseudocolored in green), and an Alox15 antibody (with secondary conjugated to AF555, red). Nuclei were stained with DAPI (blue). Representative images of lung tissue from one mouse per group show that only APM + HDM treated mice (bottom) had cells that stain positive for both markers (white arrows, overlay) (A). Alox15 positive cells that were not monocytes were present in FA + HDM treated lungs (yellow arrows). A close-up of an Alox15 positive monocyte from the APM + HDM treated group shows double staining (B). Slides were imaged using a Zeiss LSM 700 Confocal Microscope at 20× (A) and 63× (B).
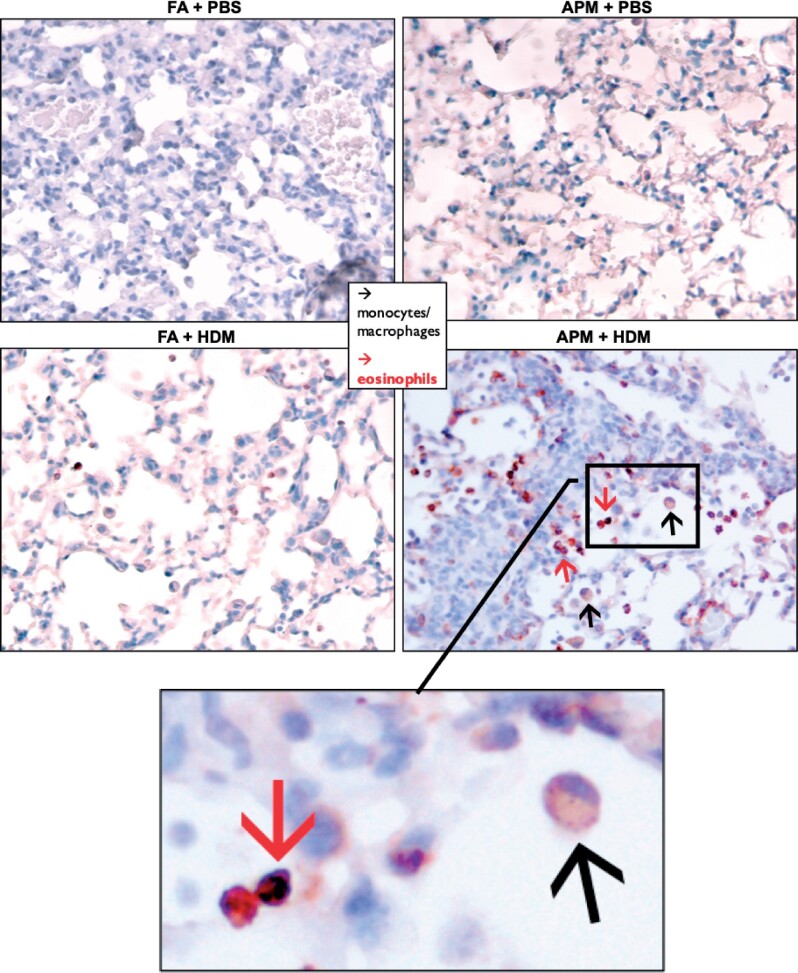
To investigate what other types of cells express Alox15, we next performed immunohistochemistry staining. We found that only lung tissue from the APM + HDM treated mice demonstrated positive Alox15 staining of both monocytes and eosinophils (Figure 5). We observed Alox15 positive staining of eosinophils in the HDM only group (no APM), but this appeared to be less than in APM + HDM treated tissue and there were no observable Alox15 positive monocytes. We also note the expression of Alox15 in airway epithelial cells of the allergen-treated groups (FA + HDM and APM + HDM). This is consistent with reports that allergens such as OVA increase the expression of Alox15 in human airway epithelial cells (Kuperman et al., 2005). Other than constitutively expressed Alox15 in epithelial cells, which were expected and observed, there was little evidence of Alox15 staining in the nonallergen groups (filtered air + PBS and APM + PBS).
Figure 5.
Positive Alox15 expression in lung tissue from APM + HDM exposed mice. Lung tissue was harvest after BAL collection and stained with Alox15 antibody then counterstained with Mayer’s hematoxylin. Representative images of tissue show that APM + HDM treated mice (bottom right) had more eosinophils (red arrows) and monocytes (black arrows) positive for Alox15 than in other groups.
APM Did Not Further Increase Eosinophilic Inflammation
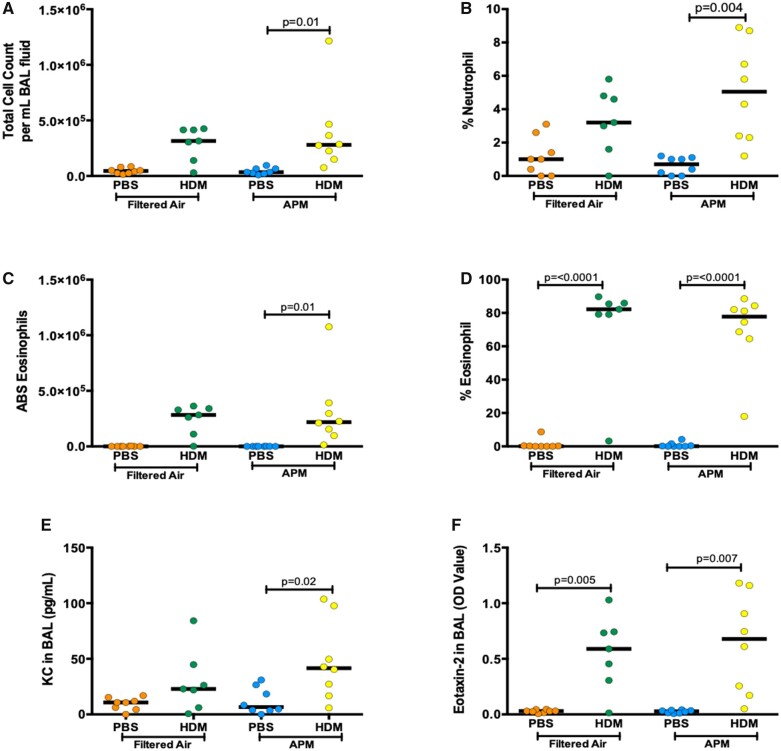
In the parallel set of mice that were treated alongside the sequencing group, we evaluated 2 major markers of DRLD: inflammation and AHR. First, we investigated the inflammatory response in BAL fluid after APM and allergen exposures. HDM treatment significantly increased total cell count, percent neutrophils, and absolute and percent eosinophils in BAL of APM pre-exposed mice (p = .01, p = .004, p = .01, and p < .0001, respectively, Figs 6A–D). In filtered air controls, HDM significantly increased the percent of eosinophils in BAL (p < .0001, Figure 6D). APM exposure alone did not induce neutrophilic or eosinophilic inflammation. We next measured levels of KC (the mouse ortholog of human IL-8) and Eotaxin-2 in cell-free BAL fluid, which are neutrophil and eosinophil chemokines, respectively. APM pre-exposure followed by allergen challenge increased KC and Eotaxin-2 production (p = .02 and p = .007, respectively, Figs 6E and 6F). There is no significant difference in filtered air versus APM without allergen in any of these parameters. This is likely due to the long recovery between the termination of APM and the initiation of the allergic asthma model. Because neutrophils generally clear within 24 h, it is likely that the inflammatory cells produced in response to APM alone had already resolved. HDM did not induce significant production of total cell count, percent neutrophils, absolute numbers of eosinophils, and KC levels compared to controls after filtered air exposure. In all figures, if there is no reported p-value, the comparison is not significant.
Figure 6A.
APM pre-exposure enhances inflammation and AHR after the HDM challenge. AHR was assessed 24 h after the last challenge (n = 8/group). BAL was collected 24 h after the last challenge from mice after airway physiology assessment. APM pre-exposure followed by allergen (APM vs APM + HDM) challenge significantly increased total cell count (A), absolute eosinophil numbers (B), % eosinophils (C), Eotaxin-2 protein (D), and KC protein (E). Data are presented with bars indicating the median. APM + HDM exposure increased AHR compared with filtered air + PBS control AHR in a dose-dependent manner (G). APM pre-exposure followed by allergen challenge significantly increased AHR at the highest Mch concentration (H). Data presented as means + SEM (G) or bars to represent median (H). p-Values represent a one-way ANOVA and p-value of less than .05 was considered significant. An outlier (determined via the outlier test) was removed from the FA + HDM group at the 50 mg/ml dose. A two-way ANOVA was performed to compare filtered air versus APM versus HDM versus APM + HDM: HDM = 0.0003, APM = 0.54, Interaction = 0.52 (A); HDM = 0.0001, APM = 0.39, Interaction = 0.1 (B); HDM = 0.0002, APM = 0.61, Interaction = 0.61 (C); HDM = 0.0001, APM = 0.86, Interaction = 0.91 (D); HDM = 0.003, APM = 0.22, Interaction = 0.35 (E); HDM = 0.0001, APM = 0.69, Interaction = 0.67 (E); HDM = 0.0027, APM = 0.058, Interaction = 0.20 (H).
Figure 6B.
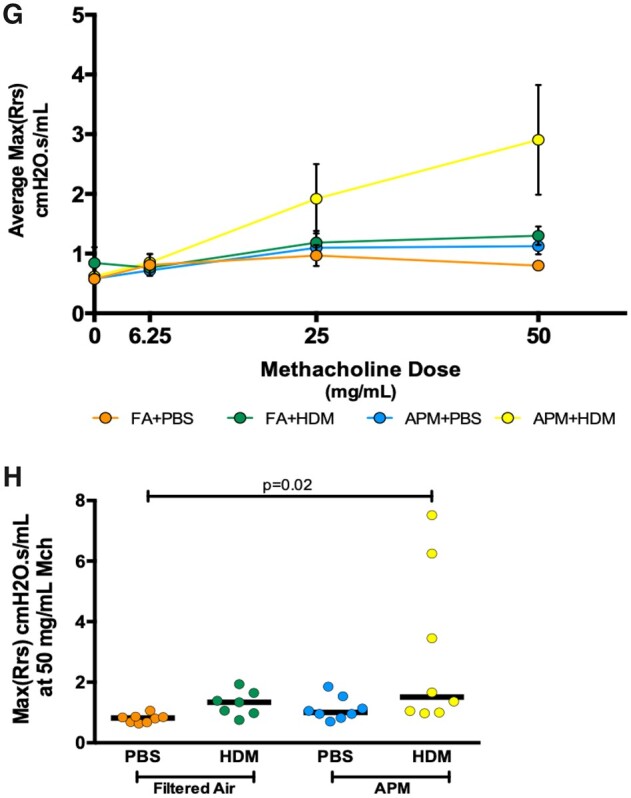
APM Pre-exposure Enhances AHR After Allergen Challenge
A key feature of both DRLD and asthma is AHR. It has been previously reported that Alox15 increases the duration of airway obstruction in asthma (Lai et al., 1990; Profita et al., 2000). Mice were exposed to increasing doses of Mch and total Rrs was evaluated as an indicator of AHR. Across all of the Mch doses, APM + HDM mice had the highest average max Rrs values (Figure 6G). At the highest dose of Mch, APM + HDM significantly enhanced AHR compared with control (filtered air + PBS) (Figure 6H). Allergen challenge (without APM) trended to have increased AHR at the 50 mg/ml Mch dose, but this was not statistically significant. Because airway physiology was assessed more than 3 weeks post-termination of APM exposure, the AHR observed was driven primarily by allergen exposure, which follows the inflammatory data.
DISCUSSION
Our findings demonstrate for the first time that exposure to APM during deployment appears to prime airways to be more responsive to common household allergens upon returning home. This priming effect may explain in part why symptoms of DRLD can persist in the postdeployment setting. We used scRNAseq of BAL cells to characterize the response of each cell after APM exposure has ended and inflammation has mostly resolved. Using this approach, we have identified a unique subset of monocytes induced only by the combination of APM and HDM. The expression of a number of genes associated with the development of allergic asthma is upregulated within this monocyte subpopulation. Taken together, we believe that Alox15 expressed by this unique population of monocytes and possibly other cell types involved in the airway response to APM + HDM (such as eosinophils) may contribute to DRLD in the postdeployment setting. DRLD represents a significant concern to the United States Armed Forces with both health and financial costs. Identification of this monocyte population as a potential contributor to disease will guide future studies, for example, with investigations focused on altering the signaling pathways or activities of these Alox15 positive cells.
The whole body APM exposures followed by allergen sensitization and challenge in mice depict an occupational exposure that military personnel may face. A key feature of some DRLD cases is AHR (Garshick et al., 2019; Morris et al., 2014). We found that mice exposed to APM followed by allergen had significantly increased AHR compared with filtered air and PBS controls. In APM (no allergen) exposed mice, there is a slight increase in AHR compared with filtered air controls, even though there are no differences in inflammation between these 2 groups. This demonstrates that even after cessation of exposure to APM and resolution of inflammation, AHR can persist to some degree. We recognize that we were unable to demonstrate a robust inflammatory and AHR phenotype as a result of APM pre-exposure. Furthermore, the increases in AHR in APM + HDM treated mice were largely driven by the allergen exposure and not the APM pretreatment. We hypothesize that inflammation resolved in the 3 weeks between the termination of APM exposure and beginning of the allergen challenge model. In future studies, we plan to initiate the allergen model before the resolution of APM inflammation and AHR can occur. This finding is significant as it suggests that, even after returning from deployment, APM-induced intracellular changes may persist. The APM-induced changes in gene expression can be seen best within the large alveolar macrophage cluster (Figure 3), where the APM-exposed cells form a cluster distinct from the filtered air controls. This reprogramming may prime airway cells for a second hit from allergen exposure.
We focused this manuscript on APM + HDM exposed mice and the monocyte cluster induced only by this treatment. This group of cells may contribute to the pathogenesis of DRLD and the impact that these exposures have on the health of our military population. We showed that only mice in this treatment group had significantly increased AHR and within this population, these cells expressed high levels of Alox15, which promotes AHR in allergic asthma. Alox15, also known as 15-lipoxygenase, is part of a group of enzymes involved in lipid peroxidation that uses arachidonic acid (AA) as a substrate (Kuhn et al., 2015). Alox15 functions by metabolizing AA to form 15-hydroxyeicosatetraenoic acid (15-HETE) (Adel et al., 2016; Snodgrass and Brune, 2019). A number of biologically active mediators are produced by lipoxygenases, including leukotrienes and lipoxins (Haeggstrom and Funk, 2011; Kuhn et al., 2016). In humans, ALOX15 has been identified as a potential therapeutic target for allergic asthma, and its expression level is reported to correlate with asthma severity (Dekker, 2016; Mabalirajan et al., 2013; Zhao et al., 2009). In a model mimicking Alox15 knockout, mice deficient in 12/15-LO (the ortholog of human ALOX15) showed decreased inflammation (indicated by differential leukocyte count) after challenge with OVA compared with WT mice (Andersson et al., 2008). In a comprehensive review of the role of the 15-lipoxygenases in human macrophages, Snodgrass and Brune (2019) reported that macrophages involved in the resolution phase of inflammation have high levels of ALOX15.
Alox15 is also known to promote AHR, and increased expression of Alox15 in this population of APM + HDM treated mice may also account for the increased AHR. Mabalirajan et al. (2013) showed that mice injected with an overexpression vector for 12/15 lipoxygenase demonstrated spontaneous AHR. Copas et al. (1982) treated human bronchial smooth muscle ex vivo with 15-HETE, the metabolite of ALOX15, and determined that the muscle contracted in response to this treatment. In a study evaluating airway physiology in atopic asthma patients, inhalation of 15-HETE in combination with an allergen promoted the early asthmatic response (including bronchoconstriction) compared with 15-HETE plus the allergen diluent (Lai et al., 1990).
Alox15 is also expressed in cell types other than monocytes—notably, eosinophils and epithelial cells, which were excluded from our single-cell analysis. The inflammatory phenotype seen in the BAL of mice who underwent the Mch challenge demonstrated that eosinophils in the APM + HDM group also stained positively for Alox15 and these mice exhibited the highest levels of eosinophilic inflammation. As eosinophils are known to express Alox15, we believe that the Alox15 from eosinophils and this rare monocyte cluster may together enhance the inflammatory response that characterizes DRLD (Esnault et al., 2013; Kuhn et al., 2015; Mashima and Okuyama, 2015; Sridhar et al., 2019; Turk et al., 1982). HDM challenge induced a robust inflammatory response compared with PBS exposure, measured by total cell count, eosinophil percentages, and eotaxin-2 production. Unfortunately, the presence of eosinophils was not reflected in the scRNAseq data. This is because granulocytes (such as eosinophils and neutrophils) are not captured by the 10× Genomics droplet-based method that we used. Chen et al. (2018) found that live (unfixed) neutrophils were not captured by the 10× system and suggest that fixation of the single cells may improve capture efficiency. Despite this, the 10× Genomics platform remains an optimal system for evaluating larger numbers of cells at a low depth to identify rare cell populations (Ziegenhain et al., 2017). Using other approaches, we were able to show that eosinophilic inflammation was induced by this allergic asthma model. Furthermore, the importance of eosinophils in allergic asthma has been well documented, so we were able to focus on novel pathways such as Alox15 expression in response to APM and allergen exposure instead.
Alox15 can also be expressed by nonimmune cells. The IHC staining showed that some airway epithelial cells also stain positively for Alox15, primarily in the HDM + PBS group. Although epithelial cells are known to express Alox15, some researchers speculate that Alox15 expression in immune cells, rather than epithelial cells, may contribute to the development of asthma and associated symptoms. Mabalirajan et al. (2013) compared lungs from normal and asthmatic patients and stained for ALOX15. The researchers found that in tissue from both normal and asthmatic patients, bronchial epithelial cells stained positively for ALOX15. However, immunohistochemical staining in the asthmatic tissue indicated the presence of ALOX15 positive inflammatory cells. There was no inflammatory cell staining for ALOX15 in the normal, nonasthmatic tissue. Our staining did reveal some constitutive expression of Alox15 in AECs, but the greatest numbers of immune cells positive for Alox15 were in the APM + HDM exposed sections. We did not see colocalization of Alox15 and monocytes/macrophages in any groups other than APM + HDM, suggesting that both treatments are required for induction of Alox15 expression. The positive Alox15 staining of inflammatory cells (monocytes, macrophages, and eosinophils) in the APM + HDM group further supports a role for Alox15 that we identified in our scRNAseq data in promoting persistent DRLD in the postdeployment setting.
In the future, a different strain of mice (eg, BALB/c) that better displays Th2 responses (compared with Th1 responses in C57BL/6 mice) may be a more suitable model for allergic asthma. This study also employed whole body aerosolized exposures, which are more physiologically relevant to the dust and sandstorms that soldiers experience abroad. Direct delivery of APM to the airways (such as oropharyngeal, intratracheal, and intranasal instillation) would ensure that more particles reached the lungs and perhaps induce a stronger and more persistent inflammatory response.
The experiments presented in this manuscript represent the first in vivo model of a post-Afghanistan deployment setting and the exposures that may be encountered. This work demonstrates, for the first time, that a small subset of monocytes may contribute to the prolonged symptoms of DRLD in the postdeployment setting. By identifying the genes upregulated in this cluster, we are able to plan future studies around therapeutic targets that may alleviate symptoms of DRLD in some U.S. military personnel.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ACKNOWLEDGMENTS
The authors would like to thank Dr Brian Wong and Dr Karen Mumy at NAMRU-Dayton, the Wright Patterson Air Force Base, for their assistance with the whole body Afghanistan Particulate Matter exposures. They would also like to thank Heather Lowers, Kate Hay-Campbell, and Dr Geoff Plumlee at the United States Geological Survey (USGS) in Lakewood, Colorado for their work characterizing the desert particulate matter. They would also like to thank Dr Elizabeth Redente for her expertise in immunofluorescence and Josh Loomis for his assistance with the confocal microscopy. Finally, they would like to thank Dr David Jackson for providing the Afghanistan PM sample used in this study.
FUNDING
This work was funded by a grant from the Department of Defense (W81XWH-16-2-0018) to Drs G.P.D., H.W.C., B.J.D., M.A.S., and C.S.R.. Supported by National Institutes of Health (NIH)/National Center for Advancing Translational Science (NCATS) Colorado CTSA Grant Number UL1 TR002535. Contents are the authors’ sole responsibility and do not necessarily represent official NIH views. K.R. is supported as an Informatics Fellow of the RNA Bioscience Initiative, University of Colorado School of Medicine.
REFERENCES
- Adel S., Karst F., Gonzalez-Lafont A., Pekarova M., Saura P., Masgrau L., Lluch J. M., Stehling S., Horn T., Kuhn H., et al. (2016). Evolutionary alteration of ALOX15 specificity optimizes the biosynthesis of antiinflammatory and proresolving lipoxins. Proc. Natl. Acad. Sci. U S A 113, E4266–E4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson C. K., Claesson H. E., Rydell-Tormanen K., Swedmark S., Hallgren A., Erjefalt J. S. (2008). Mice lacking 12/15-lipoxygenase have attenuated airway allergic inflammation and remodeling. Am. J. Respir. Cell Mol. Biol. 39, 648–656. [DOI] [PubMed] [Google Scholar]
- Berman R., Downey G. P., Dakhama A., Day B. J., Chu H. W. (2018). Afghanistan particulate matter enhances pro-inflammatory responses in IL-13-exposed human airway epithelium via TLR2 signaling. Toxicol. Sci. 166, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Cheung F., Shi R., Zhou H., Lu W., and CHI Consortium (2018). PBMC fixation and processing for Chromium single-cell RNA sequencing. J. Transl. Med. 16, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copas J. L., Borgeat P., Gardiner P. J. (1982). The actions of 5-, 12-, and 15-HETE on tracheobronchial smooth muscle. Prostaglandins Leukot. Med. 8, 105–114. [DOI] [PubMed] [Google Scholar]
- Eleftheriadis N., Dekker F. J. (2016). The role of human 15-lipoxygenase-1 in asthma. SM J. Pulm. Med. 2, 1015. [Google Scholar]
- Engelbrecht J. P., McDonald E. V., Gillies J. A., Jayanty R. K., Casuccio G., Gertler A. W. (2009). Characterizing mineral dusts and other aerosols from the Middle East–Part 2: Grab samples and re-suspensions. Inhal. Toxicol. 21, 327–336. [DOI] [PubMed] [Google Scholar]
- Esnault S., Kelly E. A., Schwantes E. A., Liu L. Y., DeLain L. P., Hauer J. A., Bochkov Y. A., Denlinger L. C., Malter J. S., Mathur S. K., et al. (2013). Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PLoS One 8, e67560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshick E., Abraham J. H., Baird C. P., Ciminera P., Downey G. P., Falvo M. J., Hart J. E., Jackson D. A., Jerrett M., Kuschner W., et al. (2019). Respiratory health after military service in southwest Asia and Afghanistan. An Official American Thoracic Society Workshop Report. Ann. Am. Thorac. Soc. 16, e1–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeggstrom J. Z., Funk C. D. (2011). Lipoxygenase and leukotriene pathways: Biochemistry, biology, and roles in disease. Chem. Rev. 111, 5866–5898. [DOI] [PubMed] [Google Scholar]
- Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F., et al. (2018). Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107.e17. [DOI] [PubMed] [Google Scholar]
- Herbert C., Siegle J. S., Shadie A. M., Nikolaysen S., Garthwaite L., Hansbro N. G., Foster P. S., Kumar R. K. (2013). Development of asthmatic inflammation in mice following early-life exposure to ambient environmental particulates and chronic allergen challenge. Dis. Model Mech. 6, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invernizzi G., Ruprecht A., Mazza R., Rossetti E., Sasco A., Nardini S., Boffi R. (2004). Particulate matter from tobacco versus diesel car exhaust: An educational perspective. Tob. Control 13, 219–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P. V., Silberstein L., Scadden D. T. (2014). Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krefft S. D., Wolff J., Zell-Baran L., Strand M., Gottschall E. B., Meehan R., Rose C. S. (2020). Respiratory diseases in post-9/11 military personnel following southwest Asia deployment. J. Occup. Environ. Med. 62, 337–343. [DOI] [PubMed] [Google Scholar]
- Kuhn H., Banthiya S., van Leyen K. (2015). Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta 1851, 308–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn H., Gehring T., Schroter A., Heydeck D. (2016). Cytokine-dependent expression regulation of ALOX15. J. Cytokine Biol. 1, 106. [Google Scholar]
- Kuperman D. A., Lewis C. C., Woodruff P. G., Rodriguez M. W., Yang Y. H., Dolganov G. M., Fahy J. V., Erle D. J. (2005). Dissecting asthma using focused transgenic modeling and functional genomics. J. Allergy Clin. Immunol. 116, 305–311. [DOI] [PubMed] [Google Scholar]
- Lai C. K., Polosa R., Holgate S. T. (1990). Effect of 15-(s)-hydroxyeicosatetraenoic acid on allergen-induced asthmatic responses. Am. Rev. Respir. Dis. 141, 1423–1427. [DOI] [PubMed] [Google Scholar]
- Lalonde D., Bradley M. (2015). 11 Years of Air Quality Monitoring in Afghanistan. Available at: https://military-medicine.com/article/3163-11-years-of-air-quality-monitoring-in-afghanistan.html. Accessed January 3, 2021.
- Mabalirajan U., Rehman R., Ahmad T., Kumar S., Leishangthem G. D., Singh S., Dinda A. K., Biswal S., Agrawal A., Ghosh B. (2013). 12/15-lipoxygenase expressed in non-epithelial cells causes airway epithelial injury in asthma. Sci. Rep. 3, 1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima R., Okuyama T. (2015). The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biol. 6, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnesota Pollution Control Agency. About the Air Quality Index. Available at: https://www.pca.state.mn.us/air/about-air-quality-data. Accessed January 3, 2021.
- Morris M. J., Dodson D. W., Lucero P. F., Haislip G. D., Gallup R. A., Nicholson K. L., Zacher L. L. (2014). Study of active duty military for pulmonary disease related to environmental deployment exposures (STAMPEDE). Am. J. Respir. Crit. Care Med. 190, 77–84. [DOI] [PubMed] [Google Scholar]
- Orellano P., Quaranta N., Reynoso J., Balbi B., Vasquez J. (2017). Effect of outdoor air pollution on asthma exacerbations in children and adults: Systematic review and multilevel meta-analysis. PLoS One 12, e0174050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Profita M., Sala A., Riccobono L., Paternò A., Mirabella A., Bonanno A., Guerrera D., Pace E., Bonsignore G., Bousquet J., et al. (2000). 15-Lipoxygenase expression and 15(S)-hydroxyeicoisatetraenoic acid release and reincorporation in induced sputum of asthmatic subjects. J. Allergy Clin. Immunol. 105, 711–716. [DOI] [PubMed] [Google Scholar]
- Snodgrass R. G., Brune B. (2019). Regulation and functions of 15-lipoxygenases in human macrophages. Front. Pharmacol. 10, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J. Y., Lee J. T., Park Y. H., Bell M. L. (2013). Short-term effects of air pollution on hospital admissions in Korea. Epidemiology 24, 545–554. [DOI] [PubMed] [Google Scholar]
- Sridhar S., Liu H., Pham T. H., Damera G., Newbold P. (2019). Modulation of blood inflammatory markers by benralizumab in patients with eosinophilic airway diseases. Respir. Res. 20, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner T. R., , WongB. A., , MumyK. L., , JamesR. A., , RebouletJ., , DoddD. E., , StriebichR. C., and , Mattie D. R. (2020). Toxicity and occupational exposure assessment for hydroprocessed esters and fatty acids (HEFA) alternative jet fuels. J. Toxicol. Environ. Health A. 83, 181–202. [DOI] [PubMed] [Google Scholar]
- Szema A., Mirsaidi N., Patel B., Viens L., Forsyth E., Li J., Dang S., Dukes B., Giraldo J., Kim P., et al. (2017). Proposed Iraq/Afghanistan War-Lung Injury (IAW-LI) clinical practice recommendations: National Academy of Sciences' Institute of Medicine Burn Pits Workshop. Am. J. Mens Health 11, 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szema A. M., Peters M. C., Weissinger K. M., Gagliano C. A., Chen J. J. (2010). New-onset asthma among soldiers serving in Iraq and Afghanistan. Allergy Asthma Proc. 31, 67–71. [DOI] [PubMed] [Google Scholar]
- Szema A. M., Reeder R. J., Harrington A. D., Schmidt M., Liu J., Golightly M., Rueb T., Hamidi S. A. (2014). Iraq dust is respirable, sharp, and metal-laden and induces lung inflammation with fibrosis in mice via IL-2 upregulation and depletion of regulatory T cells. J. Occup. Environ. Med. 56, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szema A. M., Salihi W., Savary K., Chen J. J. (2011). Respiratory symptoms necessitating spirometry among soldiers with Iraq/Afghanistan war lung injury. J. Occup. Environ. Med. 53, 961–965. [DOI] [PubMed] [Google Scholar]
- Turk J., Maas R. L., Brash A. R., Roberts L. J. 2nd, Oates J. A. (1982). Arachidonic acid 15-lipoxygenase products from human eosinophils. J. Biol. Chem. 257, 7068–7076. [PubMed] [Google Scholar]
- Vieth B., Parekh S., Ziegenhain C., Enard W., Hellmann I. (2019). A systematic evaluation of single cell RNA-seq analysis pipelines. Nat. Commun. 10, 4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weese C. B., Abraham J. H. (2009). Potential health implications associated with particulate matter exposure in deployed settings in southwest Asia. Inhal. Toxicol. 21, 291–296. [DOI] [PubMed] [Google Scholar]
- Zell-Baran L. M., Meehan R., Wolff J., Strand M., Krefft S. D., Gottschall E. B., Macedonia T. V., Gross J. E., Sanders O. L., Pepper G. C., et al. (2018). Military occupational specialty codes: Utility in predicting inhalation exposures in post-9/11 deployers. J. Occup. Environ. Med. 61,1036–1040. [DOI] [PubMed] [Google Scholar]
- Zhang J., Fulgar C. C., Mar T., Young D. E., Zhang Q., Bein K. J., Cui L., Castaneda A., Vogel C. F. A., Sun X., et al. (2018). TH17-induced neutrophils enhance the pulmonary allergic response following BALB/c exposure to house dust mite allergen and fine particulate matter from California and China. Toxicol. Sci. 164, 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Maskrey B., Balzar S., Chibana K., Mustovich A., Hu H., Trudeau J. B., O'Donnell V., Wenzel S. E. (2009). Interleukin-13-induced MUC5AC is regulated by 15-lipoxygenase 1 pathway in human bronchial epithelial cells. Am. J. Respir. Crit. Care Med. 179, 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. X., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W. (2017). Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643.e4. [DOI] [PubMed] [Google Scholar]