Abstract
In this review we describe the application of CRISPR tools for functional genomics screens in bacteria, with a focus on the use of interference (CRISPRi) approaches. We review recent developments in CRISPRi titration, which has enabled essential gene functional screens, and genome-scale pooled CRISPRi screens. We summarize progress toward enabling CRISPRi screens in non-model and pathogenic bacteria, including the development of new dCas9 variants. Taking into account the current state of the field, we provide a forward-looking analysis of CRISPRi strategies for determining gene function in bacteria.
Introduction
The exponential increase in bacterial genome assemblies and the growing importance of studying the full diversity of bacterial life have led to an increased focus on functional genomic approaches. By coupling genome-scale genetic perturbations with high-throughput phenotypic assays, functional genomics systematically defines gene-phenotype relationships, allowing functional inferences for genes of unknown function. Several high-throughput methods exist for perturbing gene function, including transposon-based approaches such as Tn-seq and TraDIS, knockout collections, and CRISPR approaches. These methods have unique strengths and weaknesses and are often deployed in a complementary fashion. However, advances in our understanding of CRISPR, decreases in DNA synthesis costs, and new CRISPR modalities have led to broad adoption of CRISPR for functional genomics studies across the bacterial domain.
There are many families of CRISPR systems, but most bacterial applications use the Type II-A system from Streptococcus pyogenes in which a nuclease effector protein (Cas9Spy, [1]) is targeted to a specific DNA sequence by a complementary RNA (either a crRNA:tracrRNA complex [1] or a fused single guide RNA [2]). The Cas9-sgRNA complex binds complementary DNA in a two-step process, first recognizing a short DNA sequence (NGG) called the protospacer adjacent motif (PAM), and then progressively unwinding the DNA and forming a DNA:sgRNA R-loop [3]. Once bound, Cas9 cuts DNA in a stereotypical position, enabling DNA editing [4,5]. In bacteria, more attention has been focused on using a catalytically deactivated Cas9 (dCas9) to downregulate transcription (CRISPR interference - CRISPRi [2,6]). CRISPRi blocks RNA polymerase (RNAP) elongation (Figure 1A, potentially perturbing the expression of downstream and upstream genes in the targeted operon ([7–10], reviewed in [11]). dCas9 has also been used to direct transcriptional activators to specific genomic sites (e.g. CRISPR activation - CRISPRa [6,12,13]).
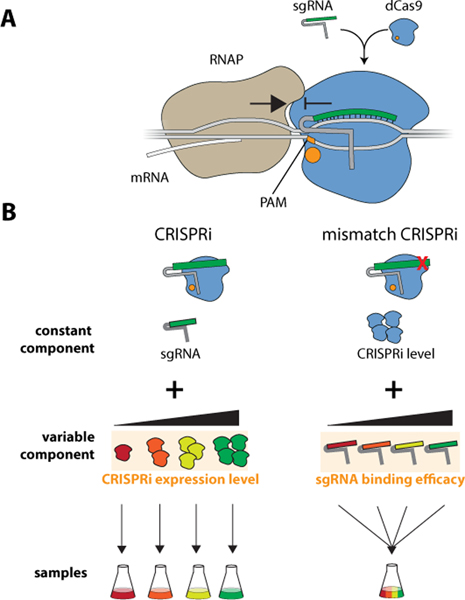
Figure 1.
A) Schematic of CRISPRi mechanism. A catalytically inactivated Cas9 (dCas9) recognizes a PAM site and unwinds the DNA allowing the base-pairing region (green) of its sgRNA to bind the complementary DNA sequence in a target gene. dCas9 acts as a roadblock to elongating RNA polymerase (RNAP) and thus represses the expression of the targeted gene. B) Adjusting repression strength in CRISPRi and mismatch CRISPRi. In regular CRISPRi (left), repression is controlled by adjusting the expression level of CRISPRi components, and each level of repression necessitates a separate experiment. In mismatch CRISPRi (right), the same effect is achieved by adjusting the binding strength of sgRNAs through the introduction of mismatches. This allows many interference levels to be queried in a single pooled sample.
CRISPR methodologies offer three major advantages. First, because CRISPR activity depends on the expression of both the sgRNA and dCas9 components, it is inducible and titratable [2,14]. This enables the creation of libraries targeting essential genes and the measurement of gene dosage effects, neither of which cannot be achieved through transposon mutagenesis or traditional knockout approaches. Second, because CRISPR sgRNA spacers contain only 20bp of unique sequence, genome-scale sgRNA libraries can be quickly synthesized, cloned, and quantified through deep sequencing using the sequence of the sgRNA spacer as the bar-code. Alternatively, small, targeted libraries can be constructed to target a specific process. Third, due to their modular nature, CRISPR systems can be engineered (e.g. different promoters, dCas9 variants, delivery systems) to function in diverse microbial species enabling similar approaches in different organisms. Limitations of CRISPRi approaches include operon level knockdown and potential dCas9 toxicity. Additionally, when inducible systems are employed, there is a temporal delay between dCas9 production and target depletion via cell division. In this review, we discuss recent developments in CRISPR technology that have buttressed these strengths and furthered the application of CRISPR systems for characterizing gene function and highlight technological and conceptual advances in functional genomics afforded by CRISPR approaches.
Titration of CRISPRi activity
The ability to control CRISPR activity has been a central motivation driving its adoption. In contrast to transposon or knockout approaches, which inactivate gene function during strain construction, most bacterial CRISPRi approaches tightly control the expression of dCas9 and/or sgRNAs to modulate knockdown, usually using inducible promoters [2,7,8,10,14–16] (Figure 1B). By constructing CRISPRi libraries in permissive conditions (dCas9/sgRNA not expressed) and performing experiments in non-permissive conditions (dCas9 and sgRNA expressed), CRISPRi enables the exploration of essential and conditionally-essential genes. All CRISPRi screens in bacteria performed to date are collated in Table 1.
Table 1 –
All CRISPRi screens in bacteria to date.
| Organism | Screen | sgRNAs | Genes | Reference |
|---|---|---|---|---|
| B. subtilis | Arrayed chemical genomics | 289 | Essential | [7] |
| Pooled growth/mismatch | 33,585 | Essential | [24] | |
| S. pneumoniae | Arrayed growth/microscopy | 384 | Essential | [15] |
| Pooled in vivo growth | 1,499 | All | [57] | |
| E. coli | Pooled growth | 92,919 | All | [8] |
| Pooled growth | ~60,000 | All | [9] | |
| Pooled growth/phage | 92,919 | All | [10] | |
| Pooled Microscopy/Growth | 235 | Cell-cycle | [42] | |
| Pooled growth | ~33,000 | All | [39] | |
| CRISPR adaptation/growth | 462,382 | All | [41] | |
| Pooled growth in multiple strains | 11,629 | Most | [58] | |
| Pooled growth/phage | ~33,000 | All | [35] | |
| Pooled growth/mismatch | 36,291 | Essential | [24] | |
| M. smegmatis | Pooled growth | 11,467 | 2,385 | [34] |
| Arrayed growth/microscopy | 272 | Essential | [19] | |
| V. natriegens | Pooled growth | 13,567 | All | [20] |
| S. mutans | Arrayed growth/microscopy | 259 | Essential | [18] |
| Synechocystis sp. | Pooled growth | 10,498 | All | [40] |
Bacillus subtilis [7] and Streptococcus pneumoniae [15] studies were the first to take advantage of this inducibility by building arrayed libraries of CRISPRi strains targeting all essential genes (identified using orthogonal genetic approaches) and probing the chemical sensitivities and cellular morphology of these strains. By identifying phenotypes shared by genes of known and unknown function, these screens revealed the roles of previously uncharacterized genes in iron-sulfur cluster biogenesis [7], peptidoglycan synthesis [15] and teichoic acid synthesis [15]. The utility of the B. subtilis CRISPRi library for determining the targets of drugs was also demonstrated both in the initial study [7] and in subsequent trials [17]. More recently, the Burne lab [18] targeted the essential genes of Streptococcus mutans, a pathogen associated with dental caries, using dCas9 from S. mutans itself. By characterizing the growth and morphology of their library, they identified the function of a poorly annotated essential gene and dissected a novel virulence determinant. Likewise, the Warner lab [19] applied CRISPRi (using dCas9 from Streptococcus thermophilus - dCas9Sth [16]) to the essential genes of Mycobacterium smegmatis, a close relative of the human pathogen M. tuberculosis. Using high-throughput microscopy, they determined the function of an uncharacterized essential gene.
Titrating CRISPR activity via inducible promoters allowed the probing of essential gene function, however testing multiple knockdown levels has remained difficult. In contrast, programming CRISPRi activity level into sgRNA sequences allows quantification of the fitness impact of multiple knockdown levels in a single pooled experiment. Early studies leveraged the decreased activity of sgRNAs designed to pair with the template (non-coding) strand [7,8,10,20], (likely because those complexes are more easily surpassed by RNAP [21]) to perform initial analyses of gene-specific susceptibility to knockdown. Control of sgRNA activity by varying the extent of complementarity between sgRNAs and their targets has also been demonstrated [22]. More recently, the Gross lab demonstrated precise control of sgRNA activity by introducing single, targeted mismatches into the sgRNA (Figure 1B, mismatch-CRISPRi [23,24]), likely by affecting DNA binding kinetics [25]. Mismatch-CRISPRi functions across species, allowing the first cross-species comparison of essential gene expression-fitness relationships. The Bikard lab [26] recently identified additional sequence determinants of sgRNA activity. Because sgRNA sequences are constrained by the genome of interest, this method does not allow precise engineering of sgRNA efficacy but does enable the design of high-activity sgRNA libraries.
A different type of gene perturbation is achieved with CRISPRa, which uses dCas9 to position a transcriptional activator adjacent to a gene and activate its expression [6,12,13]. Despite its promise, this technique remains of limited utility for high-throughput approaches due to variable efficacy and strict targeting requirements. Recent work from the Wang lab has identified and engineered a dCas9-linked activation with relaxed spacing requirements, potentially relaxing targeting requirements [27]. While no CRISPRa genetic screens have been reported in bacteria, several [28,29] have been performed in eukaryotic systems, where differential responses to knockdown and activation allow stringent identification of genes with specific phenotypes. This is an exciting prospect for bacterial functional genomics, especially because CRISPRi/CRISPRa experiments can be performed simultaneously [12,13].
Construction of sgRNA libraries
The relative simplicity of constructing and assaying libraries is a major advantage of CRISPRi. Because sgRNA spacers targeting each gene are cloned into a single chromosomal or plasmid site, construction of arrayed and pooled CRISPR libraries is simplified compared to gene deletion libraries. Compared to transposon-based approaches with similar genomic coverage, CRISPRi libraries are more compact, which facilitates construction, handling, and sequencing. This size allows their use in situations in which Tn-seq would be bottlenecked (e.g., pathogenesis experiments). The decreasing cost of DNA synthesis has made it feasible to synthesize large sgRNA libraries, leveraging CRISPRi for larger scale assays (Figure 2).
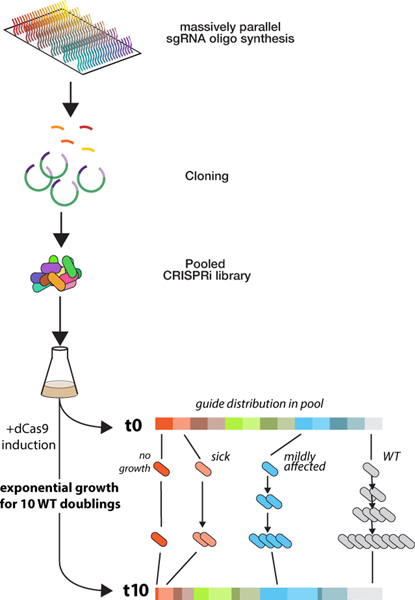
Figure 2.
Flow-chart of a pooled CRISPRi experiment. DNA oligos coding for individual sgRNAs are synthesized in bulk on a DNA chip, and cloned into the target organism as a pool. To determine the effect of downregulating a target gene through an sgRNA, the relative abundance of an sgRNA is determined via deep sequencing before and after several generations of growth. De-enrichment of an sgRNA sequence indicates a fitness defect in cells containing that sgRNA.
A major limitation of large libraries is the cloning efficiency required to maintain good representation of sgRNAs from oligo-chip to flask. Previously, this hurdle was overcome using highly efficient natural competence to introduce sgRNAs into a genomic locus (B. subtilis [24]) or by cloning sgRNAs into plasmids that can be efficiently transformed (e.g. by electroporation) [8–10,24]. Recently reported chromosomal “landing pads” [30] allow high efficiency cloning of sgRNAs directly into the genome of E. coli and other species. Integrating plasmids may allow similar functionality in other species [31–33]. Chromosomally integrated sgRNAs allow finer control of sgRNA expression and more accurate quantification of strain abundance through sequencing than plasmid-based systems. The efficiency gains from working with large, complex libraries come from assaying the collection as a pool, in many cases by quantifying the relative abundance of each strain before and after a process of selection or enrichment (e.g. during normal growth conditions to calculate relative growth rate) (Figure 2). Because substantial sequencing depth is required for accurate quantitation (for discussion, see [9,24]), these screens have benefited from the continuing reduction in sequencing costs.
The Bikard, Xing, and Warner labs performed the first sets of large library functional genomic experiments in E. coli [8–10] and M. smegmatis [34]. Using libraries of ~60,000, ~90,000, and ~12,000 elements (respectively) targeting almost all genes, these studies demonstrated the robustness of pooled screens and their utility for essential gene identification. Demonstrating the utility of these libraries to address specific biological questions, one study from the Bikard lab [10] also identified genes, including essential genes, whose knockdown reduced infection by 3 bacteriophages. A recent study expanded this approach to additional phages [35]. This highlights the ability of pooled CRISPRi approaches to determine essential gene phenotypes besides growth and paves the way for pooled CRISPRi-based chemical genomic screens, analogous to previously reported transposon studies [36]. These studies also revealed toxicity caused by high dCas9 expression and the presence of specific sgRNA seed sequences [8], similar to reports in E. coli [37] and other organisms [16,38]. These effects should be considered and addressed by evaluating large numbers of non-targeting sgRNAs to control for any non-specific phenotypes. The Church lab applied CRISPRi to Vibrio natriegens [20]. Using a library of ~14,000 sgRNAs targeting all genes, they identified its essential genes and refined metabolic gene annotations. More recently, the Arkin lab built a library of ~33,000 sgRNAs targeting all genes and almost all putative sRNAs in E. coli and screened it at various intervals post-induction [39]. These time-resolved measurements of fitness revealed gene function-specific transient responses and novel gene phenotypes. Pooled CRISPRi approaches have also been successfully applied to optimize bacteria for industrial uses [40].
The Tavazoie lab performed a large CRISPRi screen using a different approach. Rather than commercial synthesis of sgRNAs, they used the natural adaptation machinery of the S. pyogenes CRISPR-Cas system to produce sgRNAs targeting E. coli. These crRNA systems appear to function analogously to sgRNAs, but further work is required to characterize their utility [41]. Finally, recent proof-of-principle work from the Elf lab [42] demonstrated a novel method, DuMPLING, which allows cellular morphology to be assayed in a pooled setting. Scaling this concept to entire genomes and even communities is an exciting prospect.
Application of CRISPRi to diverse organisms
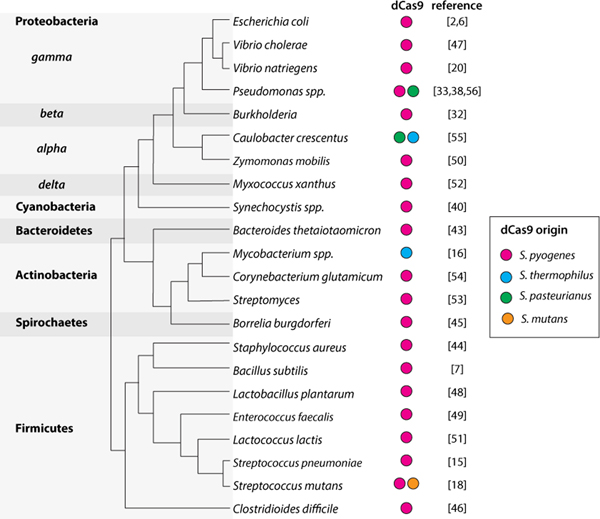
A final key advantage of CRISPRi is its applicability to diverse organisms, including many that lack genetic tools. The recognition of the importance of specific bacteria in the microbiome and environment has driven the development of techniques for working with non-model organisms, including CRISPRi approaches. In the last few years, CRISPRi has been demonstrated in many bacteria, including human associated bacteria (Figure 3). In some species, such as Bacteroidetes [43], Staphylococcus aureus [44], Borrelia burgdorferi [45], Burkholderia [32], Clostridioides difficile [46], Vibrio cholerae [47], Lactobacillus plantarum [48], Enterococcus faecalis [49], Zymomonas mobilis [50], Lactococcus lactis [51], Myxococcus xanthus [52], Streptomyces [53] and Corynebacterium glutamicum [54], it is possible to use the canonical S. pyogenes dCas9 (dCas9Spy). In other species, specialized dCas9 proteins must be used. For example, dCas9Sth is used in Mycobacterium species [16], while either dCas9Sth or S. pasteurianus dCas9 (dCas9Spa) can be used in Caulobacter crescentus [55]. Moreover, although dCas9Spy functions in Pseudomonas species, dCas9Spa was shown to be more effective in some strains [33,38,56]. A recently developed system, Mobile-CRISPRi, enables modular engineering of CRISPRi components and delivery systems, with demonstrated efficacy in ESKAPE pathogens [31]. Importantly, several studies have demonstrated that CRISPRi is maintained and functions when microbiome strains or pathogens are in the host [18,38,43], suggesting that pooled CRISPRi screens can be performed in vivo to identify genes involved in commensalism and pathogenesis. Indeed, this approach has already been applied to S. pneumoniae to successfully identify in vivo essential genes for targeted drug development [57].
Figure 3.
Bacteria in which CRISPRi has been successfully established, and origin of the dCas9 systems used.
An example of the kinds of insights that can be gained by applying CRISPRi to multiple bacterial strains can be found in the recent work from the Bikard lab [58], in which a library of 11,629 sgRNAs targeting conserved E. coli genes was introduced into 18 ecologically distinct strains. Both the fitness effects of gene knockdown and the subset of essential genes varied across strains and conditions, highlighting previously unappreciated differences within a single species. Performing CRISPRi screens in pathogenic bacteria will identify novel points of vulnerability in these species, such as the rhamnose-glucose polysaccharide synthesis pathway identified in S. mutans [18], informing antibiotic development. Cross species comparisons will enable deeper evolutionary comparisons of essential gene functions [24].
Future directions
When we last reviewed CRISPR methods in bacteria [59], no high-throughput CRISPRi screens had been performed. This has been rectified in the intervening years with a veritable explosion of high-throughput, high-dimensional CRISPRi screens of essential genes and whole genomes. These studies have revealed not only novel gene functions, but also novel connections between pathways.
Despite the inherent modularity and portability of CRISPRi approaches, almost all high-throughput screens were performed in model systems. Considering the breadth of biologically and industrially relevant organisms in which CRISPRi has been demonstrated (reviewed above), and the expanding toolkit of sgRNA design software, cloning, and sequencing approaches, CRISPRi screens in diverse bacteria (e.g. S. mutans [18], M. smegmatis [19]) may become increasingly common. In contrast to studies in well-characterized bacteria, CRISPRi screens in non-model organisms may generate numerous novel functional annotations, which will undoubtedly propagate to related species. Importantly, comparative functional genomics analyses across strains/species will allow us to study the functional conservation of genes. For example, comparison of E. coli and B. subtilis essential gene expression fitness curves has already revealed shared constraints on essential gene expression [24].
Just as CRISPR based screens have expanded from model systems to non-model organisms, CRISPR based techniques are expanding into fully fledged screens. In particular, continued engineering of dCas9 variants [12,13,60] with relaxed PAM requirements and the discovery of new transcriptional activators [27] may enable the first CRISPRa screens in bacteria. Such screens will reveal new biology. Advances in sequencing and synthesis may also drive new CRISPRi methodologies. In particular, the decreasing price and increasing quality of paired-end sequencing strategies allows the sequencing of two proximally encoded sgRNAs. Since CRISPRi can effectively target numerous genes simultaneously, it allows the quantification of fitness in strains depleted for two genes. Double knockout analysis in the yeast Sacchromyces cerevisiae furthered the understanding of its genetics by uncovering redundancy and revealing its network of genetic interactions [61]. Likewise, applying this technique to bacteria will expand our understanding of their gene networks.
Supplementary Material
Highlights.
Bacterial CRISPR approaches are used for gene editing, repression, and activation.
CRISPRi, which blocks transcription of targeted genes, is the main bacterial modality.
Chemical genomics and titratable CRISPRi enable studies of essential gene functions.
Pooled, genome-wide CRISPRi screens identify novel gene functions.
CRISPRi has been demonstrated in many non-model bacteria.
Acknowledgements
We would like to thank Jason Peters and members of the Gross lab for critically reading the manuscript and for their helpful comments. C.A.G. is supported by the National Institutes of Health (R35 GM118061) and the Innovative Genomics Institute, UC Berkeley.
References
- 1.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E: A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA: Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang F, Doudna JA: CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 2017, 46:505–529. [DOI] [PubMed] [Google Scholar]
- 4.Tong Y, Whitford CM, Robertsen HL, Blin K, Jørgensen TS, Klitgaard AK, Gren T, Jiang X, Weber T, Lee SY: Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proc Natl Acad Sci U S A 2019, 116:20366–20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, La Russa M, Qi LS: CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem 2016, 85:227–264. [DOI] [PubMed] [Google Scholar]
- 6.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA: Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 2013, 41:7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo B-M, Marta E, et al. : A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell 2016, 165:1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui L, Vigouroux A, Rousset F, Varet H, Khanna V, Bikard D: A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat Commun 2018, 9:1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, Guan C, Guo J, Liu B, Wu Y, Xie Z, Zhang C, Xing X-H: Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat Commun 2018, 9:2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rousset F, Cui L, Siouve E, Becavin C, Depardieu F, Bikard D: Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genet 2018, 14:e1007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vigouroux A, Bikard D: CRISPR tools to control gene expression in bacteria. Microbiol Mol Biol Rev 2020, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong C, Fontana J, Patel A, Carothers JM, Zalatan JG: Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat Commun 2018, 9:2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana J, Dong C, Kiattisewee C, Chavali VP, Tickman BI, Carothers JM, Zalatan JG: Effective CRISPRa-mediated control of gene expression in bacteria must overcome strict target site requirements. Nat Commun 2020, 11:1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontana J, Dong C, Ham JY, Zalatan JG, Carothers JM: Regulated Expression of sgRNAs Tunes CRISPRi in E. coli. Biotechnol J 2018, 13:e1800069. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Gallay C, Kjos M, Domenech A, Slager J, van Kessel SP, Knoops K, Sorg RA, Zhang J-R, Veening J-W: High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 2017, 13:931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, et al. : Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol 2017, 2:16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin JK 2nd, Sheehan JP, Bratton BP, Moore GM, Mateus A, Li SH-J, Kim H, Rabinowitz JD, Typas A, Savitski MM, et al. : A dual-mechanism antibiotic kills Gram-negative bacteria and avoids drug resistance. Cell 2020, 181:1518–1532.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shields RC, Walker AR, Maricic N, Chakraborty B, Underhill SAM, Burne RA: Repurposing the Streptococcus mutans CRISPR-Cas9 system to understand essential gene function. PLoS Pathog 2020, 16:e1008344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wet TJ, Winkler KR, Mhlanga MM, Mizrahi V, Warner DF: Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential Mycobacterial genes. bioRxiv 2020, doi: 10.1101/2020.03.20.000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HH, Ostrov N, Wong BG, Gold MA, Khalil AS, Church GM: Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat Microbiol 2019, 4:1105–1113. [DOI] [PubMed] [Google Scholar]
- 21.Widom JR, Rai V, Rohlman CE, Walter NG: Versatile transcription control based on reversible dCas9 binding. RNA 2019, 25:1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vigouroux A, Oldewurtel E, Cui L, Bikard D, van Teeffelen S: Tuning dCas9’s ability to block transcription enables robust, noiseless knockdown of bacterial genes. Mol Syst Biol 2018, 14:e7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jost M, Santos DA, Saunders RA, Horlbeck MA, Hawkins JS, Scaria SM, Norman TM, Hussmann JA, Liem CR, Gross CA, et al. : Titrating gene expression using libraries of systematically attenuated CRISPR guide RNAs. Nat Biotechnol 2020, 38:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins JS, Silvis MR, Koo B-M, Peters JM, Osadnik H, Jost M, Hearne CC, Weissman JS, Todor H, Gross CA: Mismatch-CRISPRi Reveals the Co-varying Expression-Fitness Relationships of Essential Genes in Escherichia coli and Bacillus subtilis. Cell Syst 2020, doi: 10.1016/j.cels.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle EA, Andreasson JOL, Chircus LM, Sternberg SH, Wu MJ, Guegler CK, Doudna JA, Greenleaf WJ: High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci U S A 2017, 114:5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvo-Villamañán A, Ng JW, Planel R, Ménager H, Chen A, Cui L, Bikard D: On-target activity predictions enable improved CRISPR-dCas9 screens in bacteria. Nucleic Acids Res 2020, 48:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho H-I, Fang JR, Cheung J, Wang HH: Programmable CRISPR-Cas transcriptional activation in bacteria. Mol Syst Biol 2020, 16:e9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Rajan SS, Friedrich MJ, Lan G, Zou X, Ponstingl H, Garyfallos DA, Liu P, Bradley A, Metzakopian E: Genome-scale CRISPRa screen identifies novel factors for cellular reprogramming. Stem Cell Reports 2019, 12:757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, Rai A, Cho MY, Stern JJ, Prota AE, et al. : Combined CRISPRi/a-based chemical genetic screens reveal that rigosertib is a microtubule-destabilizing agent. Mol Cell 2017, 68:210–223.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urtecho G, Insigne K, Tripp AD, Brinck M, Lubock NB: Genome-wide Functional Characterization of Escherichia coli Promoters and Regulatory Elements Responsible for their Function. bioRxiv 2020, [Google Scholar]
- 31.Peters JM, Koo B-M, Patino R, Heussler GE, Hearne CC, Qu J, Inclan YF, Hawkins JS, Lu CHS, Silvis MR, et al. : Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat Microbiol 2019, 4:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogan AM, Rahman ASMZ, Lightly TJ, Cardona ST: A broad-host-range CRISPRi toolkit for silencing gene expression in Burkholderia. ACS Synth Biol 2019, 8:2372–2384. [DOI] [PubMed] [Google Scholar]
- 33.Tan SZ, Reisch CR, Prather KLJ: A robust CRISPR interference gene repression system in Pseudomonas. J Bacteriol 2018, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Wet TJ, Gobe I, Mhlanga MM, Warner DF: CRISPRi-Seq for the identification and characterisation of essential Mycobacterial genes and Transcriptional Units. bioRxiv 2018, doi: 10.1101/358275. [DOI] [Google Scholar]
- 35.Mutalik VK, Adler BA, Rishi HS, Piya D, Zhong C, Koskella B, Kutter EM, Calendar R, Novichkov PS, Price MN, et al. : High-throughput mapping of the phage resistance landscape in E. coli. PLoS Biol 2020, 18:e3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price MN, Wetmore KM, Jordan Waters R, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, et al. : Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 2018, 557:503–509. [DOI] [PubMed] [Google Scholar]
- 37.Cho S, Choe D, Lee E, Kim SC, Palsson B, Cho B-K: High-Level dCas9 Expression Induces Abnormal Cell Morphology in Escherichia coli. ACS Synth Biol 2018, 7:1085–1094. [DOI] [PubMed] [Google Scholar]
- 38.Qu J, Prasad NK, Yu MA, Chen S, Lyden A, Herrera N, Silvis MR, Crawford E, Looney MR, Peters JM, et al. : Modulating pathogenesis with Mobile-CRISPRi. J Bacteriol 2019, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rishi HS, Toro E, Liu H, Wang X, Qi LS, Arkin AP: Systematic genome-wide querying of coding and non-coding functional elements in E. coli using CRISPRi. bioRxiv 2020, doi: 10.1101/2020.03.04.975888. [DOI] [Google Scholar]
- 40.Yao L, Shabestary K, Björk SM, Asplund-Samuelsson J, Joensson HN, Jahn M, Hudson EP: Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nat Commun 2020, 11:1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang W, Oikonomou P, Tavazoie S: Comprehensive genome-wide perturbations via CRISPR adaptation reveal complex genetics of antibiotic sensitivity. Cell 2020, 180:1002–1017.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camsund D, Lawson MJ, Larsson J, Jones D, Zikrin S, Fange D, Elf J: Time-resolved imaging-based CRISPRi screening. Nat Methods 2020, 17:86–92. [DOI] [PubMed] [Google Scholar]
- 43.Mimee M, Tucker AC, Voigt CA, Lu TK: Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the Murine gut Microbiota. Cell Syst 2015, 1:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato’o Y, Hisatsune J, Yu L, Sakuma T, Yamamoto T, Sugai M: Tailor-made gene silencing of Staphylococcus aureus clinical isolates by CRISPR interference. PLoS One 2018, 13:e0185987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takacs CN, Scott M, Chang Y, Kloos ZA, Irnov I, Rosa PA, Liu J, Jacobs-Wagner C: A CRISPR interference platform for selective downregulation of gene expression in Borrelia burgdorferi. 2020, doi: 10.1101/2020.10.12.335471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müh U, Pannullo AG, Weiss DS, Ellermeier CD: A xylose-inducible expression system and a CRISPR interference Plasmid for targeted knockdown of gene expression in Clostridioides difficile. J Bacteriol 2019, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caro F, Place NM, Mekalanos JJ: Analysis of lipoprotein transport depletion in Vibrio cholerae using CRISPRi. Proc Natl Acad Sci U S A 2019, 116:17013–17022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myrbråten IS, Wiull K, Salehian Z, Håvarstein LS, Straume D, Mathiesen G, Kjos M: CRISPR interference for rapid knockdown of essential cell cycle genes in Lactobacillus plantarum. mSphere 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afonina I, Ong J, Chua J, Lu T, Kline KA: Multiplex CRISPRi system enables the study of stage-specific biofilm genetic requirements in Enterococcus faecalis. MBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banta AB, Enright AL, Siletti C, Peters JM: A high-efficacy CRISPRi system for gene function discovery in Zymomonas mobilis. Appl Environ Microbiol 2020, doi: 10.1128/AEM.01621-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berlec A, Škrlec K, Kocjan J, Olenic M, Štrukelj B: Single plasmid systems for inducible dual protein expression and for CRISPR-Cas9/CRISPRi gene regulation in lactic acid bacterium Lactococcus lactis. Sci Rep 2018, 8:1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng R, Wang Y, Feng W-W, Yue X-J, Chen J-H, Hu X-Z, Li Z-F, Sheng D-H, Zhang Y-M, Li Y-Z: CRISPR/dCas9-mediated transcriptional improvement of the biosynthetic gene cluster for the epothilone production in Myxococcus xanthus. Microb Cell Fact 2018, 17:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong Y, Whitford CM, Blin K, Jørgensen TS, Weber T, Lee SY: CRISPR-Cas9, CRISPRi and CRISPR-BEST-mediated genetic manipulation in streptomycetes. Nat Protoc 2020, 15:2470–2502. [DOI] [PubMed] [Google Scholar]
- 54.Cleto S, Jensen JV, Wendisch VF, Lu TK: Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth Biol 2016, 5:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzzo M, Castro LK, Reisch CR, Guo MS, Laub MT: A CRISPR interference system for efficient and rapid gene knockdown in Caulobacter crescentus. MBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun J, Wang Q, Jiang Y, Wen Z, Yang L, Wu J, Yang S: Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microb Cell Fact 2018, 17:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Kimmey JM, de Bakker V, Nizet V, Veening J-W: Exploration of bacterial bottlenecks and Streptococcus pneumoniae pathogenesis by CRISPRi-seq. bioRxiv 2020, doi: 10.1101/2020.04.22.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rousset F, Caballero JC, Piastra-Facon F, Fernández-Rodríguez J, Clermont O, Denamur E, Rocha EPC, Bikard D: The impact of genetic diversity on gene essentiality within the E. coli species. bioRxiv 2020, doi: 10.1101/2020.05.25.114553. [DOI] [Google Scholar]
- 59.Peters JM, Silvis MR, Zhao D, Hawkins JS, Gross CA, Qi LS: Bacterial CRISPR: accomplishments and prospects. Curr Opin Microbiol 2015, 27:121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, et al. : Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. : A global genetic interaction network maps a wiring diagram of cellular function. Science 2016, 353:aaf1420–aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.