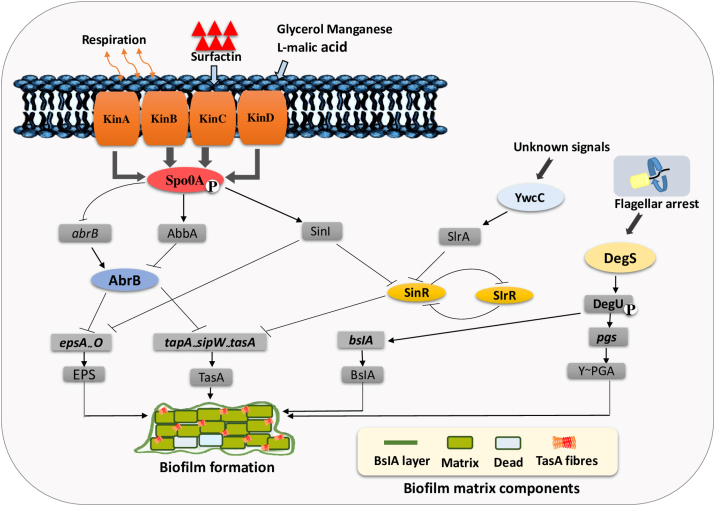
Fig. 2.
The regulatory network of B. subtilis biofilm formation. Several subnetworks are intertwined to trigger (arrows) or repress (T-bars) the expression of matrix genes involved in biofilm formation. A complex of four kinases (KinA, KinB, KinC, KinD) are activated in response to certain environmental stresses (such as temperature, high iron, oxygen concentration, hydrodynamic effects and food matrix composition). KinA and KinB are activated in response to impaired respiration in cytoplasm, KinC is activated via surfactin production and KinD is activated by glycerol, manganese and l-malic acid production. These sensor kinases initiate the activation of Spo0A through phosphorylation mediating the regulatory pathway for the expression of several matrix genes. Spo0Ã P govern an antirepressor SinI under SinR that derepresses the expression of matrix genes. In addition, SinR represses the regulatory gene slrR. SlrR binds to SinR and forms a heterodimeric complex, which leads to the transcription of the epsA-O and tapA-SipW-tasA operons, resulting in biofilm formation. Similar to SinI, the slrA gene is a paralogous antirepressor for SinR and is repressed by YwcC. Another subnetwork DegU on phosphorylation trigger the transcription of bslA indirectly, which encodes the BslA protein that construct hydrophobic coat over the biofilm. DegU trigger the activation of pgs operon directly that encodes enzymes that catalyze the synthesis of γ-PGA, which enhances biofilm robustness, for more detail see Ref. [13].