Abstract
The rapid development of mRNA-based vaccines against the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) led to the design of accelerated vaccination schedules that have been extremely effective in naive individuals. While a two-dose immunization regimen with the BNT162b2 vaccine has been demonstrated to provide a 95% efficacy in naive individuals, the effects of the second vaccine dose in individuals who have previously recovered from natural SARS-CoV-2 infection has not been investigated in detail. In this study, we characterize SARS-CoV-2 spike-specific humoral and cellular immunity in naive and previously infected individuals during and after two doses of BNT162b2 vaccination. Our results demonstrate that, while the second dose increases both the humoral and cellular immunity in naive individuals, COVID-19 recovered individuals reach their peak of immunity after the first dose. These results suggests that a second dose, according to the current standard regimen of vaccination, may be not necessary in individuals previously infected with SARS-CoV-2.
Keywords: COVID-19, T-cell immunity, BNT162b2 vaccine, SARS-CoV-2
Graphical abstract
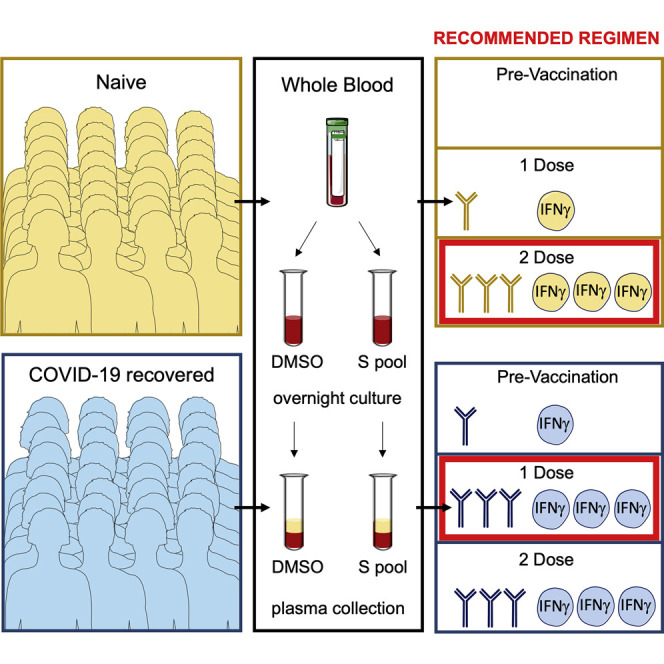
Scarce SARS-CoV-2 vaccine supplies are influencing vaccination policies in some countries. Lozano-Ojalvo et al. report that subjects with previous exposure to SARS-CoV-2 mount powerful immune responses after the first BNT162b2 vaccine dose, suggesting single vaccination regimens in COVID-19 recovered individuals.
Introduction
The BNT162b2 mRNA coronavirus disease 2019 (COVID-19) vaccine was authorized for emergency use by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) in December 2020 (Krammer, 2020). The vaccination strategy for the BNT162b2 vaccine involves an accelerated two-dose vaccination regimen administered 21 days apart, which has been demonstrated to induce a spike-specific humoral and cellular immunity associated with a 95% efficacy in naive individuals (Polack et al., 2020). However, in individuals with prior exposure to SARS-CoV-2, the utility of the second dose has been challenged. While robust spike-specific antibodies and T cells are induced by a single dose vaccination in SARS-CoV-2 seropositive individuals (Krammer et al., 2021; Prendecki et al., 2021), the second vaccination dose does not appear to further increase the spike-specific humoral response in COVID-19 recovered individuals (Samanovic et al., 2021).
The effects of the second dose of mRNA vaccine on the spike-specific T cell response has just started to be investigated in both naive and in SARS-CoV-2 pre-exposed individuals. Understanding the spike-specific T cell response is critical, as protection from disease severity and infection are likely to be dependent on the coordinated activation of both the humoral and cellular arms of adaptive immunity (McMahan et al., 2021). However, the complexity of monitoring the T cell magnitude and function has so far prevented the measurement of the immune cellular parameters in a robust number of vaccinated individuals during the full vaccination schedule. To address this problem, we investigated spike-SARS-CoV-2 antigen-specific T cell responses in naive and COVID-19 recovered individuals during full BNT162b2 mRNA vaccination.
Results
We first investigated the spike-specific T cell response by a spectral flow cytometric analysis using peripheral blood mononuclear cells (PBMCs) stimulated in vitro with DMSO or SARS-CoV-2 spike peptide pool (Le Bert et al., 2021) in naive and COVID-19 recovered individuals before and after vaccination (20 days after the second dose). We observed that spike-specific (CD154+) memory (CD45RA−CCR7−) CD4+ T cells producing both interferon (IFN)-γ and interleukin (IL)-2 were present in COVID-19 recovered individuals, as well as in naive subjects 20 days after vaccination (Figures S1A and S1B). Functional phenotyping of these cells revealed that spike-specific IFN-γ- and IL-2-secreting CD4+ T cells are mainly represented by T helper (Th)1 (CCR6−CCR4−CXCR3+) cells (Figure S1C).
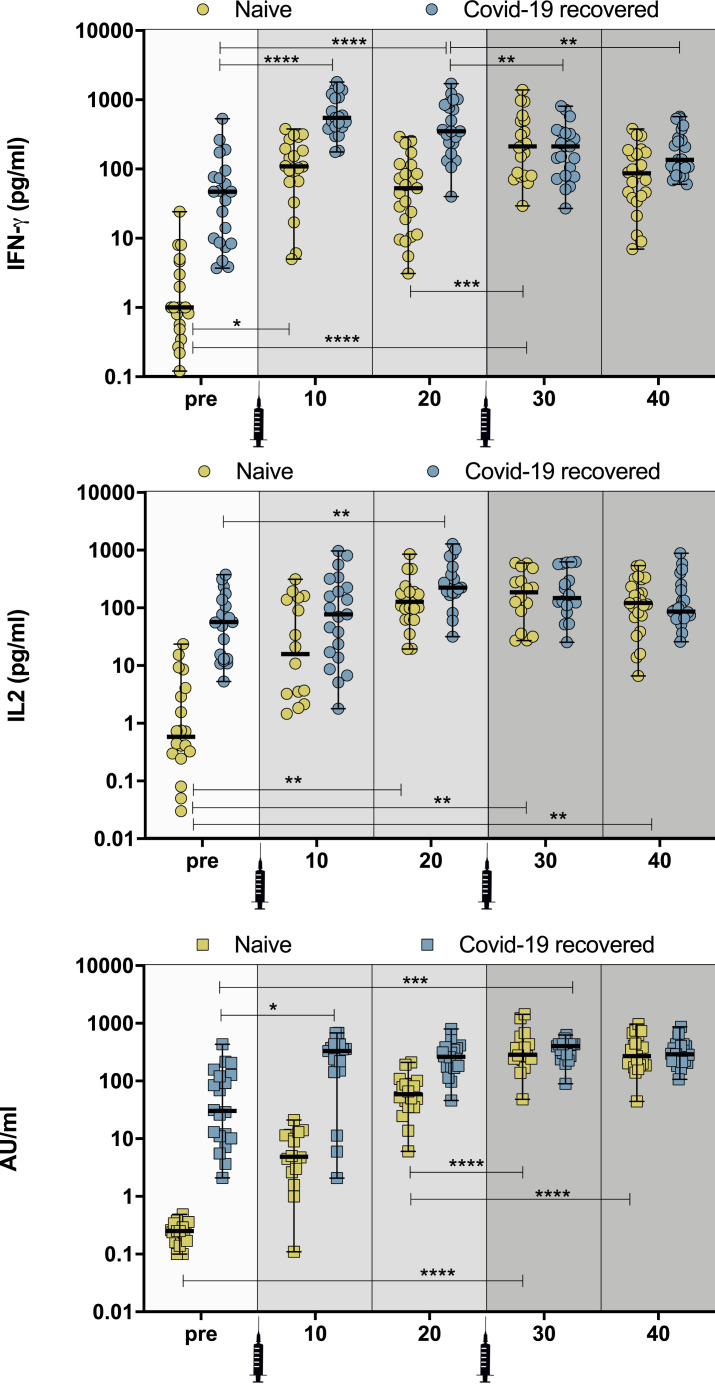
We next analyzed the dynamic changes of functional spike-specific T cell response by measuring the quantity of IFN-γ and IL-2 present in whole blood stimulated with a spike peptide pool (Le Bert et al., 2021). In contrast to more complex and detailed flow cytometry-based analyses that require larger amounts of blood, this approach requires only 1 mL of blood, which facilitates multiple longitudinal tests in a single individual and reliable quantified SARS-CoV-2-specific T cells in SARS-CoV-2-infected individuals (Le Bert et al., 2021) and vaccine recipients (Kalimuddin et al., 2021). Figure 1 shows the results obtained in 92 individuals with and without previous documented SARS-CoV-2 infection (47 naive; mean age 39.9 years [range, 20–62]; female 78%) and in 45 PCR/antigen test-confirmed SARS-CoV-2 recovered individuals (39% at 6–9 months after infection, 26% at 3–6 months, 35% at 1–3 months) (mean age 44.3 years [range, 20–76]; female 76%) (Table 1 ), tested at multiple time points (i.e., prior to and 10 and 20 days after the first and second dose of vaccine).
Figure 1.
Development of humoral and cellular immunity after BNT162b2 vaccination
(A and B) Quantification of IFN-γ (Α) and IL-2 production (B) in naive and COVID-19 recovered individuals (pre-vaccination [pre], 10 and 20 days after the first vaccine dose [10 and 20, respectively], and 10 and 20 days after the second vaccine dose [30 and 40, respectively]) after overnight stimulation of whole blood with SARS-CoV-2 peptide pools. IFN-γ and IL-2 concentration levels were determined using ELLA single plex cartridges (n = 92 individuals; 47 naive and 45 COVID-19 recovered).
(C) Kinetics of SARS-CoV-2 spike-specific IgG serum levels in naive and COVID-19 recovered individuals measured by ACCESS SARS-CoV-2 CLIA. For IgG, IFN-γ, and IL-2 determination, the group means method was used to compare pre-treatment versus post-treatment time points and also among post-treatment time points. All individual time points were measured using technical triplicates. A two-sample t test assuming unpaired populations for pre- and post-treatment was performed for each of the markers, separately for the COVID-19 recovered and naive cohorts. The p value for the test statistic was set to the 0.05 level of significance, and the Benjamini-Hochberg (BH) method was used for multiple testing correction.
Table 1.
Characteristics of the study cohort
| Naive | COVID-19 recovered | ||
|---|---|---|---|
| No. of individuals | 47 | 45 | |
| Age (mean, range) | 39.9, 20–62 | 44.3, 20–76 | |
| Female (%) | 78% | 76% | |
| Months after infection (%) | N/A | 6–9 (39%), 3–6 (26%), 1–3 (35%) | |
| IFN-γ (median, pg/mL) | Pre-vaccination | 1.0 (n = 20) | 46.9 (n = 21) |
| dose 1, 10 days (10 days) | 109.5 (n = 20) | 547.8 (n = 21) | |
| dose 1, 20 days (20 days) | 52.9 (n = 23) | 351.3 (n = 21) | |
| dose 2, 10 days (30 days) | 211.8 (n = 20) | 212 (n = 21) | |
| dose 2, 20 days (40 days) | 87.0 (n = 23) | 136.1 (n = 23) | |
| IL-2 (median, pg/mL) | Pre-vaccination | 0.5 (n = 20) | 56.9 (n = 21) |
| dose 1, 10 days (10 days) | 15.8 (n = 16) | 76.9 (n = 21) | |
| dose 1, 20 days (20 days) | 127.0 (n = 22) | 223.5 (n = 20) | |
| dose 2, 10 days (30 days) | 187.0 (n = 16) | 148.5 (n = 18) | |
| dose 2, 20 days (40 days) | 121.5 (n = 24) | 86.4 (n = 23) | |
| SARS-CoV-2 spike-specific IgG (arbitrary units [AU]/mL) | Pre-vaccination | 0.3 (n = 19) | 30.2 (n = 20) |
| dose 1, 10 days (10 days) | 4.9 (n = 16) | 332.0 (n = 21) | |
| dose 1, 20 days (20 days) | 58.9 (n = 18) | 262.8 (n = 20) | |
| dose 2, 10 days (30 days) | 284.4 (n = 16) | 402.4 (n = 20) | |
| dose 2, 20 days (40 days) | 269.7 (n = 21) | 291.5 (n = 23) |
N/A, not applicable.
Prior to vaccination, the spike peptide pool induced in whole blood a higher IFN-γ production in COVID-19 recovered individuals than in naive subjects (median pre-vaccination in naive: 1.0 pg/mL [n = 20] and in COVID-19 recovered: 46.9 pg/mL [n = 21]) (Figure 1A). We observed a similar trend in spike-specific IL-2 production (median pre-vaccination in naive: 0.5 pg/mL [n = 20] and COVID-19 recovered: 56.9 pg/mL [n = 21]) (Figure 1B; Table 1). Evaluation of the spike-specific T cell response 10 days after the first dose indicates that COVID-19 recovered individuals mount a stronger IFN-γ and IL-2 response in comparison with naive subjects (median day 10 after first vaccine dose in naive: 109.5 pg/mL [n = 20] and in COVID-19 recovered: 547.8 pg/mL [n = 21] for IFN-γ; naive: 15.85 pg/mL [n = 16] and COVID-19 recovered: 76.9 pg/mL [n = 21] for IL-2) (Figures 1A and 1B). COVID-19 recovered individuals maintain their T cell immunity on day 20 after the first vaccine dose, while IFN-γ response in naive individuals decreases, although it is not statistically significant (median day 20 after first vaccine dose in naive: 52.9 pg/mL [n = 23] and in COVID-19 recovered: 351.3 pg/mL [n = 21]). A similar trend in spike-specific IL-2 production was observed, although no decrease was seen in naive individuals (median day 20 after first vaccine dose in naive: 127.0 pg/mL [n = 22] and in COVID-19 recovered: 223.5 pg/mL [n = 20]). These results indicate that individuals with pre-existing immunity exert a more potent and sustained T cell response to SARS-CoV-2 spike after the first dose of the vaccine, consistent with recent investigations (Tauzin et al., 2021; Painter et al., 2021).
We next studied the effects of the second dose of the vaccine. Sampling on day 10 after the second dose confirmed the beneficial effects of the recall vaccine in naive individuals who increase their IFN-γ and IL-2 (median day 10 after the second vaccine dose in naive: 211.8 pg/mL [n = 20] for IFN-γ and 187.0 pg/mL [n = 16] for IL-2). On the contrary, COVID-19 recovered individuals did not increase their IFN-γ or IL-2 production on day 10 after the second vaccine dose (median day 10 after the second vaccine dose in COVID-19 recovered: 212.0 pg/mL [n = 21] for IFN-γ and 148.5 pg/mL [n = 18] for IL-2). These findings indicate that, while naive subjects significantly increase their immunity against SARS-CoV-2 spike protein after the second dose of the vaccine, COVID-19 recovered individuals do not seem to benefit from the standard regimen for COVID-19 vaccination. Interestingly, we observed that, while IFN-γ secretion in COVID-19 recovered individuals decreases on day 20 after the second dose, IL-2 levels are not decreased (median day 20 after the first vaccine dose in naive: 87.0 pg/mL [n = 23] and in COVID-19 recovered: 136.1 pg/mL [n = 23] for IFN-γ; naive: 121.5 pg/mL [n = 24] and COVID-19 recovered: 86.4 pg/mL [n = 23] for IL-2), which suggests that IL-2 secretion by spike-specific Th1 cells might represent a better biomarker to measure T cell immunity in vaccinated individuals (Figures 1A and 1B; Table 1).
Our analysis of the humoral response depicts a similar trend when measuring SARS-CoV-2 spike-specific immunoglobulin (Ig)G levels, which indicates that naive individuals achieve significant concentrations of IgG antibodies after the second vaccine dose (Figure 1C). This suggests that protection is achieved following the standard two-dose regimen for COVID-19 vaccination in naive individuals, consistent with a recent report (Ebinger et al., 2021). On the contrary, COVID-19 recovered individuals reach the peak of the response on day 10 after the first dose, which is consistent with the cellular response shown in Figures 1A and 1B. We also observed a better correlation between IL-2 and IgG in comparison to IFN-γ and IgG (Figure S2), which supports the potential use of IL-2 as biomarker for cellular immunity.
Discussion
From our studies of the humoral and cellular spike-specific immunity in recipients of the BNT162b2 mRNA COVID-19 vaccine, we conclude that a single dose of the vaccine elicits the generation of specific antibodies and a T cell immune response in naive and COVID-19 recovered individuals, consistent with recently published data (Prendecki et al., 2021; Tauzin et al., 2021). In naive subjects, the T cell response is boosted in conjunction with spike-specific IgG levels after the second dose. On the contrary, the second vaccine dose not increase the spike-specific T cell response in individuals with a pre-existing immunity against SARS-CoV-2 and further evidences that a single-dose vaccine is sufficient to induce protective immunity in Covid-19 recovered individuals (Reynolds et al., 2021).
Overall, our data support the vaccination scheme determined in clinical trials with prompt administration of the second dose in individuals without previous SARS-CoV-2 exposure (Kadire et al., 2021), but they suggest that in individuals with pre-existing immunity against SARS-CoV-2 the second dose of the vaccine may not be necessary. Longitudinal evaluation of the humoral and cellular immunity, especially in naive individuals with a reduced response to mRNA SARS-CoV-2 BNT162b2 (Grupper et al., 2021), may also justify the need for a third vaccination dose to boost the immune response in immunosuppressed individuals (Kamar et al., 2021). At present, there is a shortage of doses to vaccinate a large proportion of the population, and public health authorities are designing priority vaccination strategies according to age, comorbidities, and risk assessment criteria. The present data support a single vaccine dose in COVID-19 recovered individuals.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BV421 anti-human CD196 (CCR6, clone 11A9) | BD Horizon | 562515 |
| efluor450 anti-human IL17A (clone eBio64DEC17) | Invitrogen | 48-7179-42 |
| BV480 anti-human CD127 (clone HIL-7R-M2) | BD Horizon | 566101 |
| efluor506 anti-human CD14 (clone 61D3) | Invitrogen | 69-0149-42 |
| BV570 anti-human CD45RA (clone HI100) | BioLegend | 304132 |
| BV605 anti-human CD16 (clone 3G8) | BioLegend | 302040 |
| BV650 anti-human TNF-alpha (clone Mab11) | BioLegend | 502938 |
| Qdot655 anti-human CD27 (clone CLB-27/1) | Invitrogen | Q10066 |
| BV711 anti-human OX40 (clone ACT35) | BioLegend | 350030 |
| BV750 anti-human CD3 (clone SK7) | BioLegend | 344846 |
| BV785 anti-human CCR7 (clone G043H7) | BioLegend | 353230 |
| FITC anti-human CD154 (clone 24-31) | Invitrogen | 11-1548-42 |
| AF532 anti-human CD4 (clone RPA-T4) | Invitrogen | 58-0049-42 |
| PercP anti-human HLA-DR (clone L243) | BioLegend | 307628 |
| PercP-Cy5.5 anti-human CCR4 (clone 1G1) | BD PharMingen | 560726 |
| PercP-efluor710 anti-human CD8 (clone SK1) | Invitrogen | 46-0087-42 |
| PE anti-human IL2 (clone MQ1-17H12) | Invitrogen | 12-7029-42 |
| PE-efluor610 anti-human CXCR3 (clone CEW33D) | Invitrogen | 61-1839-42 |
| PE-Cy5 anti-human CD137 (clone 4B4-1) | BioLegend | 309808 |
| PE-Cy7 anti-human CD69 (clone FN50) | BioLegend | 310912 |
| APC anti-human IL4 (clone MP4-25D2) | BioLegend | 500812 |
| AF647 anti-human CXCR5 (clone RF8B2) | BD PharMingen | 558113 |
| AF700 anti-human IFN-gamma (clone B27) | BD PharMingen | 557995 |
| APC-efluor780 anti-human CD25 (clone BC96) | Invitrogen | 47-0259-42 |
| APC-Fire810 anti-human CD19 (clone HIB19) | BioLegend | 302272 |
| Biological samples | ||
| Peripheral blood mononuclear cells (PBMCs) | Patients and healthy donors | N/A |
| Specific antibodies measured in serum samples | Patients and healthy donors | N/A |
| IFN-gamma measured in whole blood samples | Patients and healthy donors | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Zombie NIR Fixable Viability Kit | BioLegend | 423106 |
| Ficoll-Paque Plus | GE Healthcare | GE17-1440-02 |
| AIM-V medium | GIBCO | 12055091 |
| Human AB serum | Gemini Bio-Products, Inc. | 100-512 |
| GolgiPlug (Protein Transport Inhibitor) | BD Biosciences | 555029 |
| SARS-CoV-2 peptide pools of 15-mers (55 peptides) | Le Bert et al., 2021 | N/A |
| PEPTIVATOR SARS-COV-2 PROT_S+, RG 60NMOL | MILTENY | 130-127-312 |
| PEPTIVATOR SARS-COV-2 PROT_S, RG 60 NMOL | MILTENY | 130-126-701 |
| PEPIVATOR SARS-COV-2 PROT_S1,RG 60 NMOL | MILTENY | 130-127-048 |
| Critical commercial assays | ||
| Access SARS-CoV-2 IgG Antibody Test | Beckman Coulter Inc | C58961 |
| Simple Plex Human IL-2 Cartridge | ProteinSimple | SPCKB-PS-000295 |
| Simple Plex Human IFN-gamma (3rd Gen) Cartridge | ProteinSimple | SPCKB-PS-002574 |
| Deposited data | ||
| Raw and analyzed data | This paper | N/A |
| Software and algorithms | ||
| Flow data were collected with SpectroFlo® Software | Cytek Biosciences | N/A |
| Flow cytometry data were analyzed on Cytobank | https://cytobank.org/ | N/A |
| Statistical analyses using Graphpad Prism (v9) | Graphpad Holdings, LLC | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jordi Ochando (jordi.ochando@mssm.edu).
Materials availability
Further information and requests for resources and reagents should be directed to Jordi Ochando (jordi.ochando@mssm.edu). This study did not generate new unique reagents.
Experimental model and subject details
Peripheral blood samples were obtained from healthy naive and COVID-19 recovered donors. Peripheral blood samples for humoral and cellular immunity antibody isolation were obtained from male and female donors, and the information of individuals are in Table 1. Age of these patients are undisclosed due to privacy policy. The study protocols for the collection of clinical specimens from individuals with and without SARS-CoV-2 infection were reviewed and approved by Hospital La Paz, Hospital 12 de Octubre, Hospital Gregorio Marañón, IIS-Fundación Jimenez Díaz, Hospital Universitario Marqués de Valdecilla-IDIVAL and Hospital Puerta de Hierro Clinical Research Ethics Committee (CEIm), and Mount Sinai Hospital Institutional Review Board (IRB). All the study participants provided their written informed consent for the collection of the samples and their subsequent analysis.
Method details
SARS-CoV-2 Peptide pools
SARS-CoV-2 peptide pools of 15-mers (55 peptides) covering 40.5% of the spike (S) protein contain most of the SARS-CoV2 spike epitope published to date (Grifoni et al., 2020) or pools of Prot S/S1/S+ from Miltenyi, were used as reported in (Le Bert et al., 2021).
Whole-blood culture with SARS-CoV-2 peptide pools
320 μL of whole blood drawn on the same day were mixed with 80 μL RPMI and stimulated with pools of SARS-CoV-2 peptides (S; 2 μg/ml) or DMSO control. After 15 hours of culture, the supernatant (plasma) was collected and stored at −80°C until quantification of cytokines.
Cytokine quantification and analysis
Cytokine concentrations in the plasma were quantified using Ella with microfluidic multiplex cartridges measuring IFN-γ and IL-2 following the manufacturer’s instructions (ProteinSimple, San Jose, California). The level of cytokines present in the plasma of DMSO controls was subtracted from the corresponding peptide pool stimulated samples.
Spike-specific IgG quantification
The ACCESS SARS-CoV-2 CLIA (Beckman Coulter Inc., California, USA) was used for semiquantitative detection of IgG directed against S protein RBD using serum obtained from venipuncture blood. Samples were tested on a UniCel Dxl 800 high-performance analyzer.
Flow cytometry
Peripheral blood mononuclear cells (PBMCs) were isolated by means of density centrifugation with Ficoll-Paque Plus (GE Healthcare, Pittsburgh, PA), frozen, and stored in liquid nitrogen until further use. For activation assays, thawed PBMCs were cultured in AIM-V medium (GIBCO, Grand Island, NY) with 2.5% AB human serum (Gemini Bio-Products, Inc., West Sacramento, CA), unstimulated (DMSO) or stimulated with SARS-CoV-2 spike (S) peptide pool (5 mg/mL) in presence of GolgiPlug (BD Biosciences, San Jose, CA) for 6 hours. Harvested PBMCs were stained for viability (Zombie NIR Fixable Viability Kit, BioLegend, San Diego, CA), washed and stained for surface markers, fixed in 4% paraformaldehyde (Electron Microscopy Services, Hatfield, PA), and treated with permeabilization buffer (eBioscience, San Diego, CA) before staining with labeled antibodies to detect intracellular CD154 and cytokines. The list of antibodies used for surface and cytoplasmic staining are shown in the Table S1. Stained cells were subsequently acquired on a 4-lasers CytekTM Aurora device (Cytek Biosciences, Fremont, CA) and flow cytometry data analyzed on Cytobank (https://cytobank.org/).
Quantification and statistical analysis
Illustrations were prepared using Graphpad Prism (v9) and Cytobank (https://cytobank.org/) and statistical analyses were conducted by Graphpad Prism (v9). For IgG, IFN-y and IL-2 determination, group means method was used to compare pre-treatment versus post-treatment time points and also among post treatment time points. Two sample t test assuming unpaired populations for pre- and post-treatment was performed for each of the markers, separately for the COVID-19 recovered and naive cohorts. The p value for the test statistic was set to 0.05 level of significance and BH method was used for multiple testing correction. Correlations (Figure S2) were estimated by nonparametric Spearman r correlation coefficient and significance was calculated by two tailed p value (indicated in each plot).
Acknowledgments
We acknowledge the technical contributions of Daniel Arroyo Sánchez, Jana Baranda, Sara Baztan-Morales, María Castillo de la Osa, Alejandra Comins-Boo, Carmen del Álamo Mayo, Sergio Gil-Manso, Sandra Gonzalez Beatriz; Hatem, Juan Irure-Ventura, Iria Miguens, Sara Muñoz Martinez, Monica Pereira, Catarina Rodrigues-Guerreiro, Mercedes Rodriguez-Garcia, and David San Segundo. We would also like to acknowledge Beckman Coulter for donating the equipment required for the determination of spike-specific IgG antibodies. Research reported in this publication was supported in part by the National Cancer Institute of the NIH (5R01HD102614-02; R01CA249204 and R01CA248984) and an ISMMS seed fund to E.G. The authors gratefully acknowledge use of the services and facilities of the Tisch Cancer Institute supported by a NCI Cancer Center Support Grant (P30 CA196521). M.S. was supported by a NCI training grant (T32CA078207). This work was supported by an ISMMS seed fund to J.O.; Instituto de Salud Carlos III (COV20-00668) to R.C.R.; the Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation (COVID-19 research call COV20/00181) co-financed by the European Development Regional Fund “A way to achieve Europe” to E.P.; the Instituto de Salud Carlos III, Spain (COV20/00170); the Government of Cantabria, Spain (2020UIC22-PUB-0019) to M.L.H.; the Instituto de Salud Carlos III (PI16CIII/00012) to P.P.; the Fondo Social Europeo e Iniciativa de Empleo Juvenil YEI (Grant PEJ2018-004557-A) to M.P.E.; and by REDInREN 016/009/009 ISCIII. This project has received funding from the European Union Horizon 2020 research and innovation programs VACCELERATE and INsTRuCT under grant agreements 101037867 and 860003.
Author contributions
Overall design of the project, E.G., A.B., and J.O.; acquisition of experimental data, all co-authors; generation of reagents and scientific inputs, all co-authors. J.O., D.L.-O., E.G., and A.B. wrote the manuscript with input from all co-authors.
Declaration of interests
A.B. declares the filing of a patent application relating to the use of peptide pools in whole blood for detection of SARS-CoV-2 T cells (pending). The remaining authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Published: August 4, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109570.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ebinger J.E., Fert-Bober J., Printsev I., Wu M., Sun N., Prostko J.C., Frias E.C., Stewart J.L., Van Eyk J.E., Braun J.G. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat. Med. 2021;27:981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupper A., Rabinowich L., Schwartz D., Schwartz I.F., Ben-Yehoyada M., Shashar M., Katchman E., Halperin T., Turner D., Goykhman Y. Reduced humoral response to mRNA SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients without prior exposure to the virus. Am. J. Transplant. 2021 doi: 10.1111/ajt.16615. Published online April 18, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadire S.R., Wachter R.M., Lurie N. Delayed second dose versus standard regimen for Covid-19 vaccination. N. Engl. J. Med. 2021;384:e28. doi: 10.1056/NEJMclde2101987. [DOI] [PubMed] [Google Scholar]
- Kalimuddin S., Tham C.Y., Qui M., de Alwis R., Sim J.X., Lim J.M., Tan H.C., Syenina A., Zhang S.L., Le Bert N. Early T cell and binding antibody responses are associated with Covid-19 RNA vaccine efficacy onset. Med (NY) 2021;2:682–688.e4. doi: 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N., Abravanel F., Marion O., Couat C., Izopet J., Del Bello A. Three doses of an mRNA Covid-19 vaccine in solid-organ transplant recipients. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2108861. Published online June 23, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Krammer F., Srivastava K., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Bielak D.A., Carreño J.M., Chernet R.L. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N. Engl. J. Med. 2021;384:1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S. Rapid induction of antigen-specific CD4+ T cells guides coordinated humoral and cellular immune responses to SARS-CoV-2 mRNA vaccination. bioRxiv. 2021 doi: 10.1101/2021.04.21.440862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., C4591001 Clinical Trial Group Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendecki M., Clarke C., Brown J., Cox A., Gleeson S., Guckian M., Randell P., Pria A.D., Lightstone L., Xu X.N. Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine. Lancet. 2021;397:1178–1181. doi: 10.1016/S0140-6736(21)00502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds C.J., Pade C., Gibbons J.M., Butler D.K., Otter A.D., Menacho K., Fontana M., Smit A., Sackville-West J.E., Cutino-Moguel T., UK COVIDsortium Immune Correlates Network. UK COVIDsortium Investigators Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021 doi: 10.1126/science.abh1282. Published online April 30, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanovic M.I., Cornelius A.R., Wilson J.P., Karmacharya T., Gray-Gaillard S.L., Allen J.R., Hyman S.W., Moritz G., Ali M., Koralov S.B. Poor antigen-specific responses to the second BNT162b2 mRNA vaccine dose in SARS-CoV-2-experienced individuals. medRxiv. 2021 doi: 10.1101/2021.02.07.21251311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussieres G., Brassard N., Laumaea A., Vezina D., Prevost J. A single BNT162b2 mRNA dose elicits antibodies with Fc-mediated effector functions and boost pre-existing humoral and T cell responses. bioRxiv. 2021 doi: 10.1101/2021.03.18.435972. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.