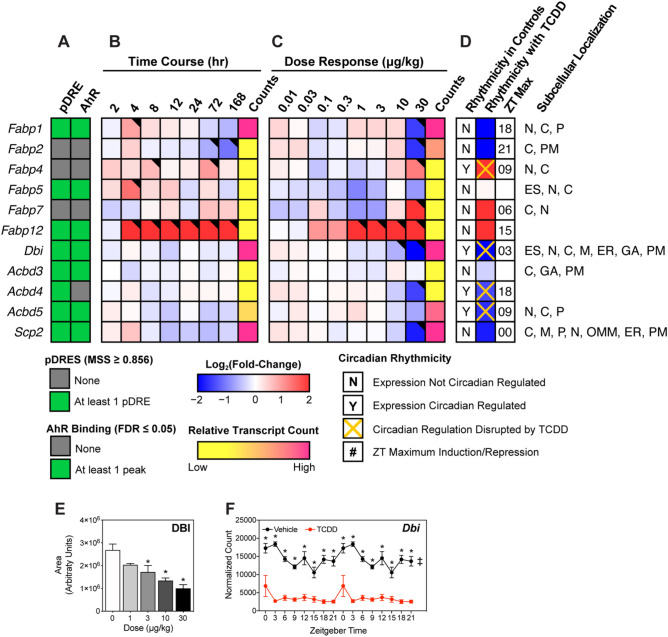
Figure 2.
Effect of TCDD on intracellular fatty acid trafficking proteins. Differential expression of genes associated with intracellular fatty acid trafficking assessed using RNA-seq. Mouse Genome Informatics (MGI) official gene symbols are used. (A) The presence of putative dioxin response elements (pDREs) and AHR genomic binding at 2 h. (B) Time-dependent gene expression following a single bolus dose of 30 μg/kg TCDD (n = 3). (C) Dose-dependent gene expression assessed following oral gavage every 4 days for 28 days with TCDD (n = 3). (D) Diurnal regulated gene expression denoted with a “Y”. An orange ‘X’ indicates abolished diurnal rhythm following oral gavage with 30 μg/kg TCDD every 4 days for 28 days. ZT indicates statistically significant (P1(t) > 0.8) time of maximum gene induction/repression. Counts represent the maximum number of raw aligned reads for any treatment group. Low counts (< 500 reads) are denoted in yellow with high counts (> 10,000) in pink. Differential gene expression with a posterior probability (P1(t)) > 0.80 is indicated by a black triangle in the top right tile corner. Protein subcellular locations were obtained from COMPARTMENTS and abbreviated as: cytosol (C), endoplasmic reticulum (ER), extracellular space (ES), Golgi apparatus (GA), mitochondrion (M), outer mitochondrial membrane (OMM), nucleus (N), peroxisome (P), and plasma membrane (PM). (E) Capillary electrophoresis was used to assess DBI protein levels in total lysate prepared from liver samples harvested between ZT0-3 (n = 3). Bar graphs denote the mean ± SEM. Statistical significance (*p ≤ 0.05) was determined using a one-way ANOVA followed by Dunnett’s post-hoc analysis. (F) Diurnal expression of Dbi was assessed by RNA-seq (n = 3). Asterisks denotes a posterior probability (P1(t) > 0.8) within the same timepoint comparing vehicle to TCDD. Diurnal rhythmicity (‡) was determined using JTK_CYCLE for each treatment. Circadian data are plotted twice along the x-axis to better visualize the gene expression rhythmicity. The heatmap was created using R (v4.0.4). Plots were created using GraphPad Prism (v8.4.3).