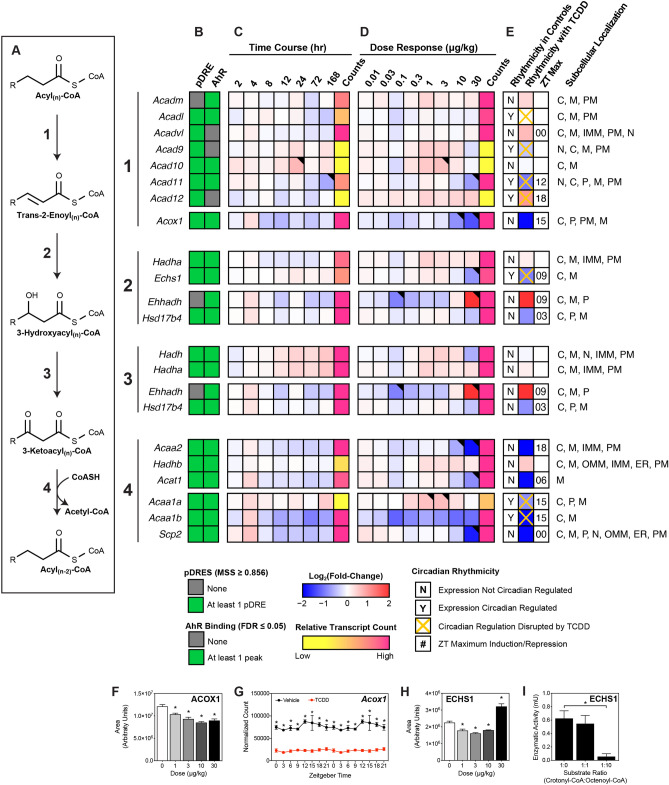
Figure 5.
Effect of TCDD on β-Oxidation of acyl-CoAs. Differential expression of genes associated with β-oxidation assessed by RNA-seq. (A) Fatty acid β-oxidation is sequentially catalyzed via acyl-CoA dehydrogenases, enoyl-CoA hydratases, hydroxyacyl-CoA dehydrogenases and thiolases. Official gene symbol designated in the MGI database are listed. (B) The presence of putative dioxin response elements (pDREs) and AHR enrichment at 2 h. (C) Time-dependent gene expression was assessed following a single bolus dose of 30 μg/kg TCDD (n = 3). (D) Dose-dependent gene expression following oral gavaged every 4 days for 28 days with TCDD (n = 3). (E) Circadian regulated genes are denoted with a “Y”. An orange ‘X’ indicates abolished diurnal rhythm following oral gavage with 30 μg/kg TCDD every 4 days for 28 days. ZT indicates statistically significant (P1(t) > 0.8) time of maximum gene induction/repression. Counts represent the maximum number of raw aligned reads for any treatment group. Low counts (< 500 reads) are denoted in yellow with high counts (> 10,000) in pink. Differential expression with a posterior probability (P1(t)) > 0.80 is indicated with a black triangle in the top right tile corner. Protein subcellular locations were obtained from COMPARTMENTS and abbreviated as: cytosol (C), mitochondrion (M), mitochondrial outer membrane (OMM), mitochondrial inner membrane (IMM), nucleus (N), peroxisome (P), and plasma membrane (PM). (F/H) Capillary electrophoresis was used to assess ACOX1 and ECHS1 protein levels in total lysate prepared from liver samples harvested between ZT0-3 (n = 3). Bar graphs denote the mean ± SEM. Statistical significance (*p ≤ 0.05) was determined using a one-way ANOVA followed by Dunnett’s post-hoc analysis. (G) Diurnal expression of Acox1 was assessed by RNA-seq (n = 3). Asterisks denotes a posterior probability (P1(t) > 0.8) within the same timepoint comparing vehicle to TCDD. Diurnal rhythmicity (‡) was determined using JTK_CYCLE for each treatment. Circadian data are plotted twice along the x-axis to better visualize the gene expression rhythmicity. (I) ECHS1 activity was assessed by monitoring the depletion of crotonyl-CoA which has an absorbance at 263 nm. Statistical significance (*p ≤ 0.05) was determined using a one-way ANOVA followed by Dunnett’s post-hoc analysis. The heatmap was created using R (v4.0.4). Plots were created using GraphPad Prism (v8.4.3). The biochemical reaction was created using Adobe Illustrator (v25.2).