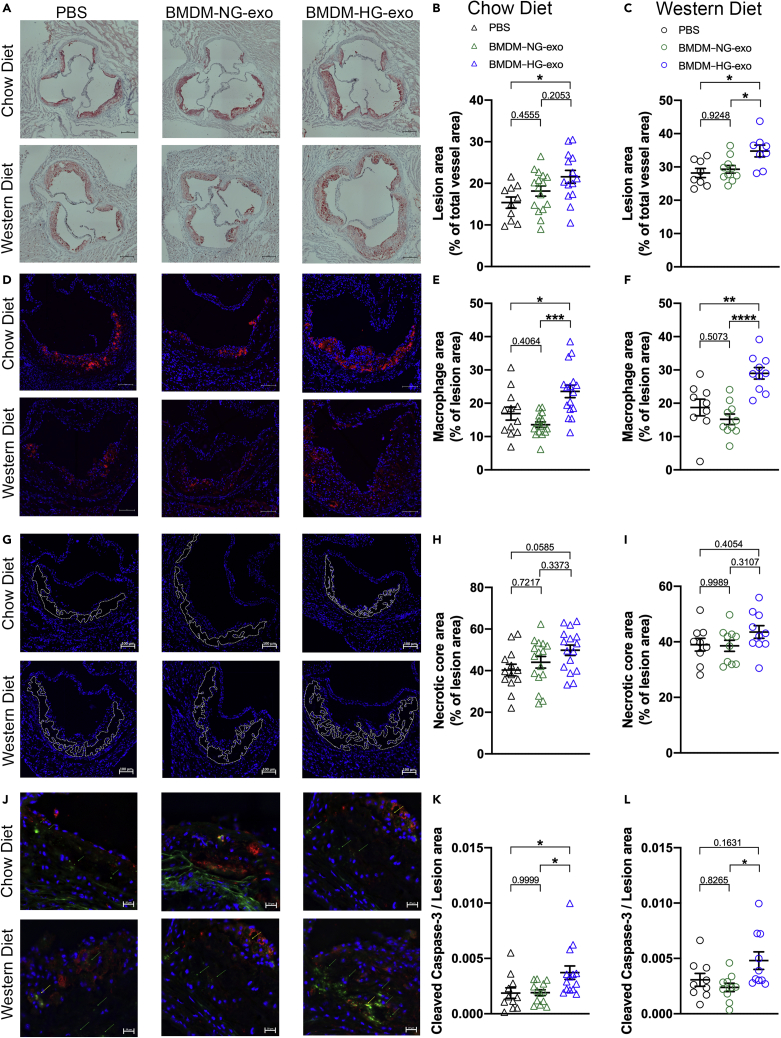
Figure 3.
BMDM–HG-exo accelerate spontaneous and diet-induced atherosclerosis in Apoe−/− mice
Apoe−/− mice were subjected to PBS, BMDM–NG-exo or BMDM–HG-exo (1×1010 particles/mice every two days) for 4 weeks.
(A and B) (A) Representative images and (B) quantification of Oil Red O (ORO) staining in Apoe−/− mice fed a chow diet. Pool of four independent experiments is shown, n = 10–16 mice in each group. Scale bars: 200μm.
(C) Quantification of ORO staining in Apoe−/− mice fed a Western diet. Pool of two independent experiments is shown, n = 8–10 mice in each group.
(D) Representative images of MOMA-2+ macrophages in the atherosclerotic plaques of aortic root areas. Scale bars: 100μm.
(E) Macrophage content analysis via immunodetection of MOMA-2 in Apoe−/− mice fed a chow diet. Pool of four independent experiments is shown, n = 12–16 mice in each group.
(F) Macrophage content analysis in Apoe−/− mice fed a Western diet. Pool of two independent experiments is shown, n = 9–10 mice in each group.
(G) Representative cross-sectional view of aortic root stained with DAPI to measure necrosis area from each group of mice. Dashed lines show the boundary of the developing necrotic core. Scale bars: 100 μm.
(H) Quantification of necrotic core area as a percent of total plaque area in Apoe−/− mice fed a chow diet. Pool of four independent experiments is shown, n = 13–16 mice in each group.
(I) Quantification of necrotic core area in Apoe−/− mice fed a Western diet. Pool of two independent experiments is shown, n = 9–10 mice in each group.
(J) Representative image of cleaved caspase-3 and MOMA-2 staining in aortic root lesions. Scale bars: 20 μm.
(K) Quantification of cleaved caspase-3 staining as a ratio of lesion area in Apoe−/− mice fed a chow diet. Pool of four independent experiments is shown, n = 11–14 mice in each group.
(L) Quantification of cleaved caspase-3 in Apoe−/− mice fed a Western diet. Pool of two independent experiments is shown, n = 9–10 mice in each group. Statistical analysis was performed using one-way ANOVA and Sidak's multiple comparisons post test. ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001. Data are represented as mean ± SEM.