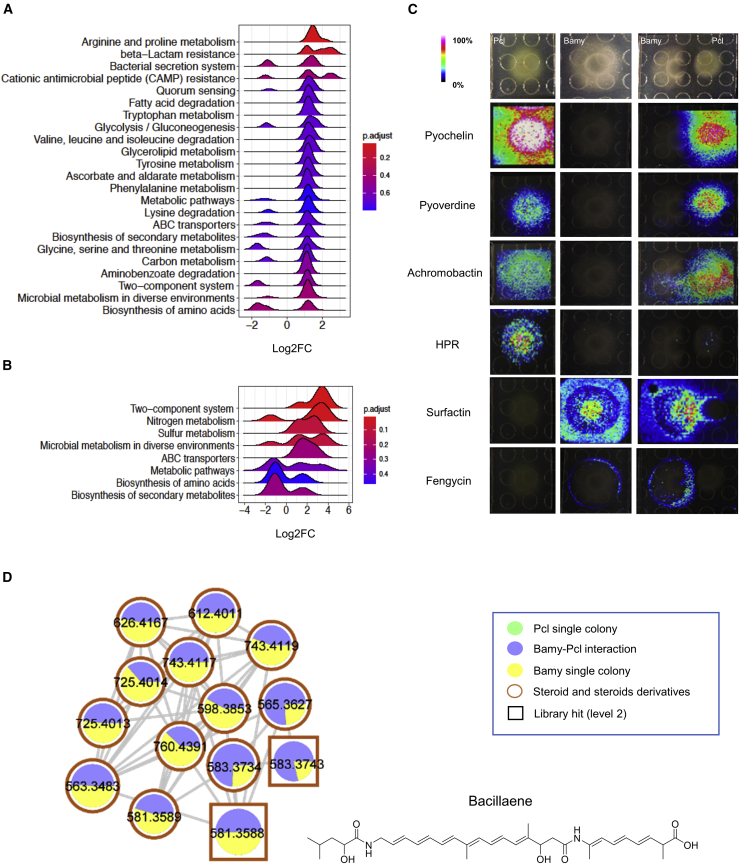
Figure 2.
Production and spatial distribution of putative antimicrobials by Pcl and Bamy
(A and B) Transcriptional analysis of overexpressed KEGG pathways in (A) Pcl and (B) Bamy after interaction for 24 h.
(C) MALDI-MSI heatmaps showing the spatial distribution of siderophores (pyoverdine, pyochelin, and achromobactin) and other secondary metabolites (HPR, surfactin, and fengycin) produced by Pcl and Bamy growing alone or in interaction.
(D) Molecular family of bacillaene detected from Bamy grown as a single colony or in the interaction with Pcl. Results were obtained in mass spectrometry analysis using LC-MS/MS and feature-based molecular networking. Each metabolite is represented by a circle, and they are connected according to the mass fragmentation patterns. The chemical structures of the annotated features are based on spectral matches in GNPS libraries representative of specific molecular families. Border colors indicate ClassyFire classification. The sizes of the compounds are directly related to their abundance in the metabolome. Squares indicate a library hit level 2 through GNPS, and circles indicate unknown compounds based on GNPS searches.