Figure 6.
Mutations in glpK provoke changes in membrane rigidity and permeability
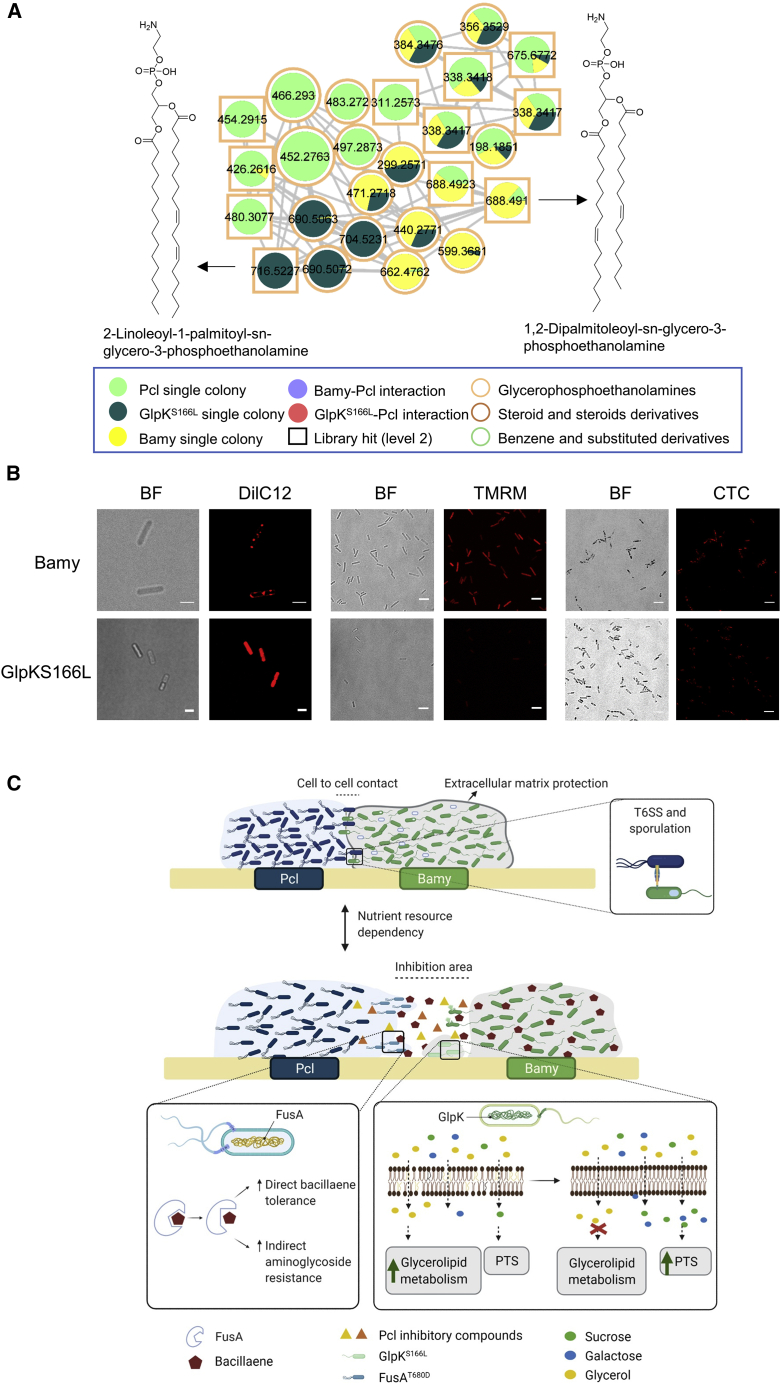
(A) Glycerophosphoethanolamine cluster, obtained by mass spectrometry analysis using LC-MS/MS and feature-based molecular networking, showing modifications in the abundance of different glycerophosphoethanolamine compounds between Pcl, Bamy, and GlpKS166L. Each metabolite is represented by a circle and they are connected according to the mass fragmentation patterns. Chemical structures of annotated features are based on spectral matches to GNPS libraries, as representative of these molecular families. Border color indicates ClassyFire classification. The sizes of the compounds are directly related to their abundance in the metabolome. Squares indicate a library hit level 2 through GNPS, while circles indicate unknown compounds based on GNPS.
(B) Membrane staining of Bamy and GlpKS166L with (left panel) DilC12 dye to analyze differences in membrane fluidity (scale bars, 2 μm), (middle panel) TMRM straining for membrane potential analysis (scale bars, 5 μm), and (right panel) CTC staining to measure respiration changes (scale bars, 5 μm). For each experiment and sample, at least three fields of view were measured.
(C) Schematic representation of the proposed interaction between Bacillus and Pseudomonas depending on critical factors, e.g., nutrients and distance. (Upper part) When bacterial populations are in contact, T6SS, sporulation, and the extracellular matrix play important roles in the interaction. (Bottom part) When populations are physically separated, the production of secondary metabolites is critical for the evolution of the interaction. Mutations in Pseudomonas fusA permit adaptation to Bacillus-produced bacillaene, thereby increasing resistance to bacillaene and aminoglycoside antibiotics. Mutations in Bacillus glpK provoke a wide array of transcriptional and metabolic changes, an increase in bacterial membrane rigidity, and a reduction of membrane potential resulting in increased antibiotic tolerance.