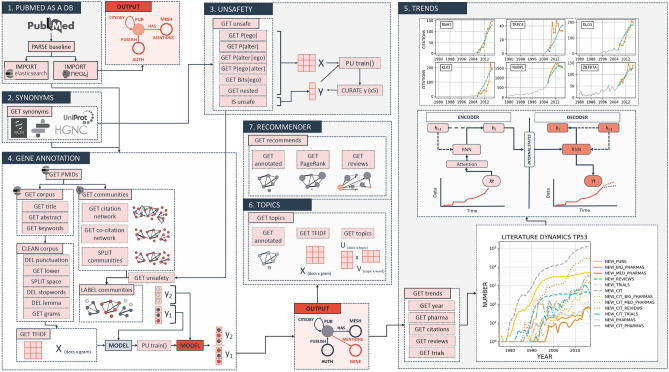
Figure 1.
Workflow. Chart summarising the process from the downloading of the data to the detection and analysis of trends in the literature. (A) Creation of a graph database with the information contained in PubMed baseline 2020. (B) Acquisition of a comprehensive collection of human coding gene names and synonyms. (C) Automatic determination of potential ambiguous (unsafe) gene names. (D) Annotation of the graph database with unambiguous gene symbols by combining co-citation network topology and binary classifiers. (E) Prediction of per-gene publication trends using RNN. When a gene has significantly more publications or citations than expected by the model it is considered to be trendy. (F) Automatic topic detection of collections of publications. We used this algorithm to quantify the evolution of topics in trendy gene publications over time. (G) A review recommender system that uses information from the citation network and topic detection to recommend the most efficient set of reviews to explore the literature.