Fig. 1.
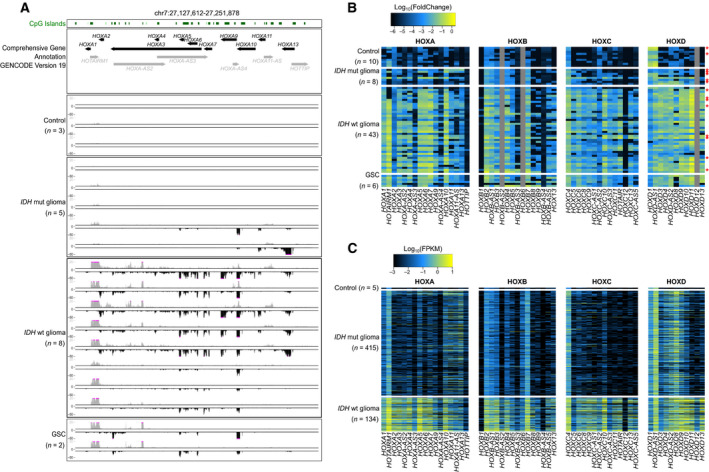
Widespread reactivation of HOX transcripts in IDHwt glioma samples. (A) Strand‐oriented RNA‐seq signals along the HOXA cluster in control (n = 3), IDHmut (n = 5) and IDHwt (n = 8) glioma and Glioma stem cells (GSC) (n = 2) samples. For each sample, sense (in black) and antisense (in gray) transcription signals are shown in the lower and upper panels, respectively. (B, C) Relative expression level of HOX coding and noncoding transcripts in control, IDHmut and IDHwt glioma, and GSC samples from our cohort analyzed by microfluidic‐based RT‐qPCR (B) and from the TCGA cohort, analyzed by RNA‐seq (C). In (B), values are the fold change relative to the geometrical mean of expression of the housekeeping genes PPIA, TBP, and HPRT1. In our cohort, expression of HOXB‐AS2, HOXB6, and HOXD12 was not analyzed. Samples analyzed both by RNA‐seq (A) and microfluidic‐based RT‐qPCR (B) are indicated with a red star in (B). FPKM, fragment per kilobase per million reads.
