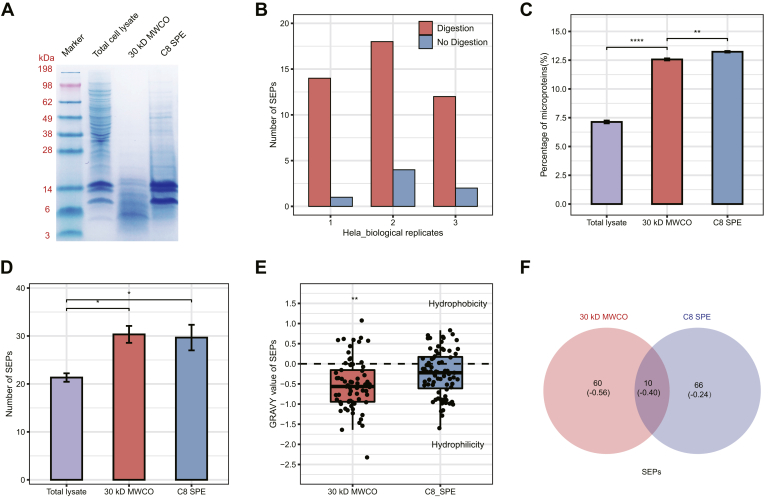
Fig. 2.
Comparison of different strategies for the identification of lncRNA-encoded SEPs.A, tricine SDS-PAGE analysis of total cell lysates, SEP fractions enriched by 30-kDa MWCO membrane and C8 SPE column from HEK293T cells are shown from left to right, respectively. B, the number of SEPs identified in HeLa cells from three biological replicates at a single MS run with (red) and without (blue) tryptic digestion. C, percentage of annotated microproteins identified in the total cell lysate and enriched fractions from HEK293T cells with three technical replicates. D, the number of SEPs identified in total cell lysate and enriched fractions from HEK293T cells with three technical replicates. E, GRAVY values of the total SEPs identified in the fractions enriched by 30-kDa MWCO membrane and C8 SPE column from HEK293T cells. F, Venn diagram of the total SEPs identified in the fractions enriched by 30-kDa membrane and C8 SPE column from HEK293T cells. (Values in parentheses show the average GRAVY values of SEPs in each region). GRAVY, grand average of hydropathicity; HEK293T, human embryonic kidney 293T; lncRNA, long noncoding RNA; MWCO, molecular weight cutoff; SEP, small ORF-encoded polypeptide; SPE, solid-phase extraction.