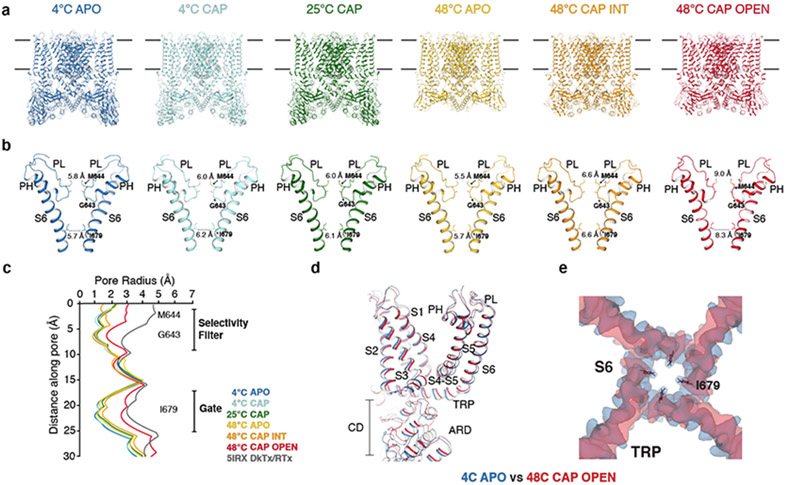
Fig. 1. Structures of the full-length TRPV1 at six conditions.
a, Structures of TRPV1 determined at 4°C (TRPV14C,APO, blue), at 4°C with capsaicin (TRPV14C,CAP, sky blue), 25°C with capsaicin (TRPV125C,CAP, green), 48°C (TRPV148C,APO, gold), 48°C with capsaicin in the intermediate state (TRPV148C,CAP,INT, orange), and 48°C with capsaicin in the open state (TRPV148C,CAP,OPEN, red). b, Comparison of the pore domain structures. Only two subunits are shown for clarity with pore loop (PL) and pore helix (PH) as indicated. Diagonal distances at the two narrowest restriction points are shown. c, Pore radii calculated using the HOLE program32 for the TRPV1 structures as color coded in panel a and DkTx/RTx-TRPV1 (PDB 5IRX, gray). d,e, Superposition of the TRPV14C,APO (blue) and the TRPV14C,CAP,OPEN (red) structures (d) and cryo-EM maps (e), highlighting global conformational changes (d) and S6 gate conformation (e), respectively.