Abstract
It was previously reported that PRR34‐AS1 was overexpressed in some solid tumors. PRR34‐AS1 promoter was shown to have a differential methylation region (DMR), and was hypomethylated in acute myeloid leukemia (AML). Therefore, the present study used real‐time quantitative PCR (RQ‐PCR) to explore the expression characteristics of PRR34‐AS1 in AML. In addition, the correlation between the expression of PRR34‐AS1 and clinical prognosis of AML was determined. The findings of this study indicated that high PRR34‐AS1 expression was bound up with shorter overall survival (OS) in AML patients (p = 0.002). Moreover, patients with high expression of PRR34‐AS1 had significantly lower complete remission (CR) rate compared with those with low expression of PRR34‐AS1 after induction chemotherapy. Furthermore, multivariate analysis confirmed that PRR34‐AS1 expression was an independent factor affecting CR in whole‐AML, non‐APL‐AML, and CN‐AML patients (p = 0.032, 0.039, and 0.036, respectively). Methylation‐specific PCR (MSP) and bisulfite sequencing PCR (BSP) were used to explore the methylation status of PRR34‐AS1. PRR34‐AS1 promoter showed a pattern of hypomethylation in AML patients compared with normal controls (p = 0.122). Notably, of whole‐AML and non‐APL‐AML patients, PRR34‐AS1 hypomethylated patients presented a significantly shorter OS than those with a hypermethylated PRR34‐AS1 (p = 0.010 and 0.037, respectively). Multivariate analysis confirmed that the hypomethylation of PRR34‐AS1 served as an independent prognostic indicator in both whole‐cohort AML and non‐APL‐AML categories (p = 0.057 and 0.018, respectively). In summary, the findings of this study showed that abnormalities in PRR34‐AS1 are associated with poor prognosis in AML. Therefore, monitoring this index may be important in the prognosis of AML and can provide information on effective chemotherapy against the disease.
Keywords: acute myeloid leukemia, DNA methylation, expression, lncRNAs, prognosis, PRR34‐AS1
In summary, the findings of this study show that high PRR34‐AS1 expression may be associated with poor chemotherapeutic efficacy and poor prognosis in AML patients. In addition, higher expression of PRR34‐AS1 was associated with the hypomethylation of its promoter, and hypomethylation of PRR34‐AS1 may affect the prognosis of AML patients.
1. INTRODUCTION
Acute myeloid leukemia (AML) presents with features of the accumulation of myeloid leukemia cells in bone marrow (BM), blood, and other tissues, The disease mainly results in poorly differentiated erythrocytes, platelets, and white blood cells (WBC) in the BM. 1 In addition, AML can occur at all ages although the incidence rate is highest in the elderly (>60 years). 2 Cytogenetic analysis has been conventionally used to study the molecular pathogenesis of leukemia for more than 50 years since the 1960s. 3 In addition, cytogenetic findings are reported to be important diagnostic and prognostic markers. 4 However, almost half of AML patients have normal karyotypes. Advances in targeted sequencing technology have led to the identification of some genetic mutations such as FLT3, NPM1, KIT, CEBPA, and TET2 in AML. 5 Most previous studies mainly focused on protein‐coding genes to explore the molecular genetic changes and identify the prognostic markers of AML. 6 However, the molecular mechanisms underlying the occurrence and development of AML have not been fully elucidated due to the high degree of heterogeneity in the disease. 7 , 8 , 9 Therefore, exploring the pathogenesis of AML is important for the development of better treatment strategies and for improving the prognosis of patients.
Abnormal regulation of long non‐coding RNAs (lncRNAs) was involved in each stage of tumor occurrence, development, and migration. 10 Moreover, genome‐wide association studies (GWAS) on tumor samples showed that numerous lncRNAs are associated with various types of cancer. 11 LncRNAs play a significant role in promoting or inhibiting the development of AML. 6 Carcinogenic lncRNAs include H19, 12 , 13 HOTAIR, 14 , 15 , 16 and PVT‐1 17 , 18 whereas tumor‐suppressing lncRNAs include NEAT1, 19 , 20 IRAIN, 21 and MEG3. 22 , 23 , 24
PRR34 antisense RNA 1 (PRR34‐AS1) was shown to be upregulated in hepatocellular carcinoma and pediatric medulloblastoma. 25 , 26 In addition, cholangiocarcinoma patients with high expression of PRR34‐AS1 were reported to have a shorter disease‐free survival (DFS). Analysis of possible mechanism showed that PRR34‐AS1 acted through the JAK‐signal transducer and activated the transcription of factors in the JAK‐STAT pathway. 27 Furthermore, PRR34‐AS1 was shown to exert its effects through the JAK‐STAT signaling pathway after total knee arthroplasty (TKA) ischemia/reperfusion (I/R) injury. 28 The differential methylation region (DMR) in the PRR34‐AS1 promoter was reported to be hypomethylated in AML. 29 However, the direct role and clinical significance of PRR34‐AS1 expression in AML have not been fully elucidated. Moreover, PRR34‐AS1 promoter methylation status and its clinical correlation with AML should be explored. Therefore, the present study sought to explore the expression and methylation characteristics of PRR34‐AS1 and their clinical significance in AML.
2. MATERIALS AND METHODS
2.1. Patients’ samples
A total of 84 newly diagnosed AML adult patients and 29 healthy controls from our hospital were enrolled in this study. Participants provided written informed consent prior to the study and ethical approval was obtained from the hospital's ethical committee. Diagnosis and classification of cases in the study were performed based on the 2016 World Health Organization (WHO) criteria. The treatment protocol is listed in Table S1.
2.2. Real‐time quantitative PCR
Bone marrow mononuclear cells (BMMNCs) were obtained using density gradient centrifugation. Total RNA was extracted from BMMCs. Reverse transcription of RNA was performed to generate cDNA, 30 following a protocol described previously. 31 Real‐time quantitative PCR (RQ‐PCR) was used to determine the expression levels of PRR34‐AS1 in the BMMNCs. The following upstream and downstream primer sequences were used to determine the expression levels of PRR34‐AS1; 5’‐GAGGCCATCTTTGGAAAGTAAA‐3’ and 5’‐AACGATGTGAGCCGAGCA‐3’, respectively. RQ‐PCR was conducted using a 20 µl reaction volume containing 20 ng of cDNA, 0.8 µM of primers, 6 µl of H2O, 10 µM of SYBR Premix TB Green (Takara), and 0.4 µM of ROX Reference Dye II (Takara). RQ‐PCR reaction conditions were as follows; 95°C for 30 s followed by 40 cycles of 95°C for 5 s, 61°C for 32 s, finally 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s. Each test included a negative control and positive control, and false positives and false negatives were excluded, respectively. ABL1 was used as the internal reference gene, and PRR34‐AS1 transcript levels in various samples were determined using the 2−ΔΔCT method.
2.3. DNA isolation, bisulfite modification, and methylation‐specific PCR
Genomic DNA was extracted from samples obtained from AML patients, AML culture cells, and normal controls using the Genomic DNA Purification Kit (Gentra). Genomic DNA was modified using the CpGenome DNA modification kit (Chemicon). Methylation‐specific PCR (MSP) was used to explore the methylation status of the PRR34‐AS1 promoter. Forward and reverse primer sequences used for methylated PRR34‐AS1 (M‐PRR34‐AS1) were 5’GGAAATGTTTAGGTCGAGGC‐3’ and 5’‐CACACATCAAAACGAAAACG‐3’, respectively. Upstream and downstream primer sequences for unmethylated PRR34‐AS1 (U‐PRR34‐AS1) were; 5’‐TATGGAAATGTTTAGGTTGAGGT‐3’ and 5’‐CACACACATCAAAACAAAAACAA‐3’, respectively. The reaction conditions were as follows; 95°C for 30 s followed by 40 cycles of 95°C for 5 s, 61°C for 32 s, 72°C for 30 s, and 78°C for 32 s. PRR34‐AS1 methylation levels were then calculated using the method.
2.4. Bisulfite sequencing PCR
Bisulfite sequencing PCR (BSP) was used to explore the density of methylation in PRR34‐AS1 and evaluate the accuracy of MSP. Notably, investigating differential methylation through BSP is a key step in the analysis of epigenetic data. 32 Upstream and downstream primer sequences for BSP were; 5’‐TTGGTATGGGAGGAGTTAAGTT‐3’ and 5’‐AAATCCCAACAACCATATACAA‐3’, respectively. The reaction conditions of BSP were pre‐denaturation (98°C for 10 s), denaturation (98°C for 10 s), annealing (61°C for 30 s), elongation (72°C for 30 s), and enzyme inactivation (72°C for 7 min). The number of cycles was set at 40. After purification and recovery, the recombinant vector was constructed using the pMD®19‐T vector (Takara), then transfected into DH5α competent cells (Vazyme Biotech Co.) for cloning. Sequences of six independent clones from each specimen were verified (BGI Gene Technology Co., Ltd.).
2.5. Gene mutation detection
LightScanner software was used to design specific primers for gene hot spots. High‐resolution melt analysis (HRMA) was used to examine mutations in N /K‐RAS, IDH1/2, DNMT3A, U2AF1, NPM1, and C‐KIT. 33 , 34 , 35 , 36 Mutation and mutation type were determined by observing the melting curve and Tm shift. Direct DNA sequencing was used to assess mutations in CEBPA and FLT3‐ITD, 37 and to verify all positive specimens.
2.6. Bioinformatics analyses
PRR34‐AS1 mRNA expression (RNA Seq V2 RSEM) and methylation (HM450) data were retrieved from a cohort of 200 AML patients in the Cancer Genome Atlas (TCGA) 5 and downloaded through the cBioPortal tool (http://www.cbioportal.org). 38 , 39 GenomicScape (http://genomicscape.com/) webserver was used for GEP analysis to further verify the relationship between the expression levels of PRR34‐AS1 and prognosis of AML. Differential methylation analysis was accomplished by the Disease Meth version 2.0 tool (http://www.bio‐bigdata.com/diseasemath/analysis.html).
2.7. Statistical analysis
Data analysis was conducted using SPSS version 22.0 software (SPSS) and GraphPad Prism 8.0. Mann–Whitney U test was used to perform comparisons between continuous variables. Comparison between the two groups of categorical variables was conducted using the Pearson's chi‐square analysis test or the Fisher exact test. Receiver operating characteristic (ROC) curve and area under the ROC curve (AUC) were used to explore differences in levels of PRR34‐AS1 methylation between AML patients and controls. Survival analysis was conducted using Kaplan–Meier survival estimates. Cox regression analysis was performed to evaluate the effect of expression and methylation of PRR34‐AS1 on the clinical outcomes of AML patients. Finally, the Spearman's rank correlation analysis was used to examine the correlation between the two groups of variables (expression and methylation of PRR34‐AS1 in AML patients). A p value <0.05 was considered to be statistically significant (bilateral).
3. RESULTS
3.1. Associations between the expression of PRR34‐AS1 and clinical as well as laboratory characteristics in AML patients
Expression levels of PRR34‐AS1 in AML patients were determined using RQ‐PCR. Analysis showed that the transcript levels of PRR34‐AS1 ranged from 0.000 to 20.339 (median 0.613) in 83 newly diagnosed AML patients. To explore the clinical characteristics of PRR34‐AS1, the cohort was group into high and low‐expression groups using the median value of the level of expression as the cutoff. The findings showed that high expression of PRR34‐AS1 was associated with higher levels of WBC (p = 0.041), platelets (p = 0.004), and older age (p < 0.001, Table 1). Moreover, there was a significant difference in the classification of karyotypes between the groups (p = 0.043). In addition, the frequency of favorable karyotypes in the high PRR34‐AS1 expression group was relatively lower compared with the level for the low‐expression group; however, the difference was not significant (14% vs. 32%, p = 0.059; Table 1).
TABLE 1.
Comparison of clinical manifestations and laboratory features between AML patients with low and high PRR34‐AS1 expression
| Patient's parameters | Low (n = 41) | High (n = 42) | p value |
|---|---|---|---|
| Sex, male/female | 20/21 | 28/14 | 0.101 |
| Median hemoglobin, g/L (range) | 77 (34–141) | 80 (49–131) | 0.600 |
| Median age, years (range) | 48 (22–84) | 60 (29–85) | <0.001 |
| Median WBC, ×109/L (range) | 6.9 (0.3–140.2) | 27.65 (0.8–528.0) | 0.041 |
| Median platelets, ×109/L (range) | 25 (3–144) | 49 (9–415) | 0.004 |
| BM blasts, % (range) | 42.5 (3–91.0) | 37 (6.5–97.5) | 0.212 |
| FAB subtypes | 0.028 | ||
| M0 | 0 (0%) | 1 (2.4%) | |
| M1 | 1 (2.4%) | 1 (2.4%) | |
| M2 | 15 (36.6%) | 15 (35.7%) | |
| M3 | 11 (26.8%) | 3 (7.1%) | |
| M4 | 5 (12.2%) | 9 (21.4%) | |
| M5 | 1 (2.4%) | 7 (16.7%) | |
| M6 | 0 (0%) | 1 (2.4%) | |
| Karyotype classification | 0.043 | ||
| Favorable | 13 (31.7%) | 6 (14.3%) | |
| Intermediate | 24 (58.5%) | 32 (76.2%) | |
| Poor | 4 (9.8%) | 1 (2.4%) | |
| No data | 0 (0%) | 3 (7.1%) | |
| Karyotype | 0.093 | ||
| Normal | 20 (48.8%) | 24 (57.1%) | |
| t(8;21) | 3 (7.3%) | 2 (4.8%) | |
| t(15;17) | 10 (24.4%) | 3 (7.1%) | |
| t(9;22) | 0 (0%) | 1 (2.4%) | |
| +8 | 0 (0%) | 2 (4.8%) | |
| −7/7q− | 1 (2.4%) | 0 (0%) | |
| complex | 3 (7.3%) | 1 (2.4%) | |
| others | 4 (9.8%) | 6 (14.3%) | |
| No data | 0 (0%) | 3 (7.1%) | |
| Gene mutation | |||
| CEBPA (+/−) | 3/32 | 4/22 | 0.689 |
| NPM1 (+/−) | 4/28 | 3/23 | >0.999 |
| FLT3‐ITD (+/−) | 3/29 | 5/21 | 0.446 |
| C‐KIT (+/−) | 2/30 | 0/26 | 0.497 |
| N/K‐RAS (+/−) | 0/24 | 2/21 | 0.234 |
| IDH1/2 (+/−) | 0/32 | 1/25 | 0.448 |
| DNMT3A (+/−) | 2/30 | 2/24 | >0.999 |
| U2AF1 (+/−) | 0/32 | 1/25 | 0.448 |
| CR (+/−) | 24/9 | 13/22 | 0.004 |
Abbreviations: BM, bone marrow; CR, complete remission; FAB, French–American–British; WBC, white blood cells.
3.2. Association between the expression of PRR34‐AS1 and efficacy of chemotherapy in AML patients
Analysis showed that patients with high expression of PRR34‐AS1 had a lower CR rate compared with those with low expression of the gene [37.1% (13/22) vs. 72.7% (24/9), p = 0.004, Table 1]. In addition, expression levels of PRR34‐AS1 were analyzed in patients with CR and those without CR after induction chemotherapy. Analysis showed that non‐CR patients had significantly higher levels of PRR34‐AS1 compared with patients with CR (p = 0.03; Figure 1A). Further, differences in clinical characteristics of AML patients with and without CR were explored. The findings showed that patients in the non‐CR group were older, had high expression levels of PRR34‐AS1 and higher levels of WBC and platelets compared with patients with CR (p = 0.010, 0.003, 0.009, and 0.004, respectively; Table 2). Moreover, the analysis showed a significant decrease in the frequency of favorable karyotypes in the non‐CR group compared with the CR group (9.7% vs. 40.5%, p = 0.005; Table 2).
FIGURE 1.
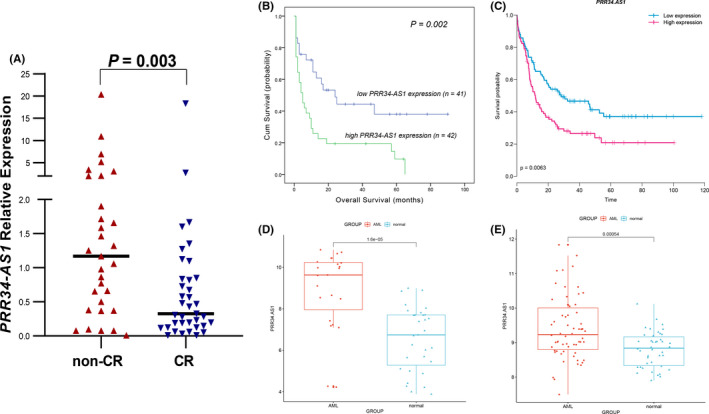
Predictive value of PRR34‐AS1 expression on CR rate and OS in AML patients. (A) Expression of PRR34‐AS1 in CR and non‐CR AML patients receiving induction therapy. (B) Overall survival (OS) of the whole cohort of AML patients. (C) Prognostic value of PRR34‐AS1 expression for OS in whole AML patients from TCGA database (GSE68833, n = 183). (D, E) PRR34‐AS1 expression level in AML patients and controls obtained from Gene Expression Omnibus (GEO)database. [D, GSE24006: AML = 23, Normal = 31; E, GSE63270: AML = 62, Normal = 42]
TABLE 2.
Comparison of clinical manifestations and laboratory features between CR and non‐CR in AML patients receiving induction therapy
| Patient's parameters | Non‐CR (n = 31) | CR (n = 37) | p value |
|---|---|---|---|
| PRR34‐AS1 expression | 1.2 (0–20.3) | 0.3 (0–18.3) | 0.003 |
| Sex, male/female | 21/10 | 18/19 | 0.143 |
| Median hemoglobin, g/L (range) | 80 (49–138) | 80 (34–131) | 0.810 |
| Median age, years (range) | 60 (22–81) | 48 (24–77) | 0.010 |
| Median WBC, ×109/L (range) | 38.7 (0.9–185.4) | 9.1 (0.8–528.0) | 0.009 |
| Median platelets, ×109/L (range) | 42 (9–415) | 32 (3–192) | 0.004 |
| BM blasts, % (range) | 37.75 (6.5–92) | 37 (3.0–97.5) | 0.143 |
| FAB subtypes | 0.038 | ||
| M0 | 1 (3.2%) | 0 (0%) | |
| M1 | 2 (6.5%) | 0 (0%) | |
| M2 | 13 (41.9%) | 15 (40.5%) | |
| M3 | 2 (6.5%) | 11 (29.7%) | |
| M4 | 8 (25.8%) | 6 (16.2%) | |
| M5 | 5 (16.1%) | 2 (5.4%) | |
| M6 | 0 (0%) | 1 (2.7%) | |
| Karyotype classification | 0.005 | ||
| Favorable | 3 (9.7%) | 15 (40.5%) | |
| Intermediate | 22 (71%) | 21 (56.8%) | |
| Poor | 4 (12.9%) | 0 (0%) | |
| No data | 2 (6.5%) | 1 (2.7%) | |
| Karyotype | 0.016 | ||
| Normal | 18 (58.1%) | 16 (43.2%) | |
| t (8;21) | 0 (0%) | 5 (13.5%) | |
| t (15;17) | 2 (6.5%) | 10 (27%) | |
| t (9;22) | 1 (3.2%) | 0 (0%) | |
| +8 | 1 (3.2%) | 1 (2.7%) | |
| −7/7q‐ | 1 (3.2%) | 0 (0%) | |
| complex | 3 (9.7%) | 0 (0%) | |
| others | 3 (9.7%) | 4 (10.8%) | |
| No data | 2 (6.5%) | 1 (2.7%) | |
| Gene mutation | |||
| CEBPA (+/‐) | 3/20 | 4/24 | >0.999 |
| NPM1 (+/‐) | 3/20 | 3/25 | >0.999 |
| FLT3‐ITD (+/‐) | 4/19 | 3/25 | 0.687 |
| C‐KIT (+/‐) | 0/23 | 2/26 | 0.495 |
| N/K‐RAS (+/‐) | 2/18 | 0/25 | 0.192 |
| IDH1/2 (+/‐) | 1/22 | 0/28 | 0.451 |
| DNMT3A (+/‐) | 2/21 | 2/26 | >0.999 |
| U2AF1 (+/‐) | 1/22 | 0/28 | 0.451 |
Abbreviations: BM, bone marrow; CR, complete remission; WBC, white blood cells.
Furthermore, logistics regression analysis further showed that PRR34‐AS1 expression could serve as an independent factor affecting CR in patients with whole AML, non‐APL‐AML, and CN‐AML (Table 3).
TABLE 3.
Univariate and multivariate analyses of variables for complete remission in whole‐cohort AML patients, non‐APL‐AML, and CN‐AML
| Variables | whole‐AML (n = 68) | non‐APL‐AML (n = 56) | CN‐AML (n = 45) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | ||||||||
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | ||
| WBC | 0.199 (0.069–0.575) | 0.003 | 0.228 (0.072–0.721) | 0.012 | 0.252 (0.079–0.800) | 0.019 | 0.196 (0.054–0.713) | 0.013 | 0.273 (0.067–1.102) | 0.068 | 1.005 (0.996–1.013) | 0.311 | |
| Age | 0.294 (0.103–0.843) | 0.023 | 0.941 (0.257–3.445) | 0.926 | 0.441 (0.141–1.383) | 0.160 | 1.455 (0.343–6.168) | 0.611 | 0.417 (0.103–1.679) | 0.218 | — | — | |
| PRR34‐AS1 expression | 0.222 (0.079–0.620) | 0.004 | 0.282 (0.089–0.895) | 0.032 | 0.272 (0.086–0.859) | 0.026 | 0.253 (0.069–0.933) | 0.039 | 0.210 (0.050–0.879) | 0.033 | 0.210 (0.050–0.879) | 0.033 | |
| Karyotype risk | 0.268 (0.102–0.706) | 0.008 | 0.351 (0.132–0.932) | 0.036 | 0.356 (0.111–1.142) | 0.083 | 0.332 (0.095–1.158) | 0.084 | — | — | — | — | |
|
CEBPA mutation |
1.111 (0.222–5.560) | 0.898 | — | — | 1.714 (0.329–8.943) | 0.522 | — | — | 0.917 (0.110–7.666) | 0.936 | — | — | |
| NPM1 mutation | 0.888 (0.145–4.401) | 0.798 | — | — | 1.200 (0.210–6.842) | 0.837 | — | — | 0.917 (0.110–7.666) | 0.936 | — | — | |
| FLT3‐ITD mutation | 0.570 (0.114–2.856) | 0.494 | — | — | 0.531 (0.085–3.310) | 0.498 | — | — | 0.556 (0.077–4.009) | 0.560 | — | — | |
| DNMT3A mutation | 0.808 (0.105–6.228) | 0.838 | — | — | 1.187 (0.150–9.408) | 0.871 | — | — | 2.000 (0.159–25.115) | 0.591 | — | — | |
Variables including age (≤60 vs. <60 years), WBC (≥30 × 109 vs.<30 × 109/L), PRR34‐AS1 expression (low vs. high), karyotype risk (favorable vs. intermediate vs. poor), and gene mutations (mutant vs. wild type).
Multivariate analysis includes variables with p < 0.200 in univariate analysis.
Abbreviations: AML, acute myeloid leukemia; CI, confidence interval; CN‐AML, cytogenetically normal AML; HR, hazard ratio; non‐APL‐AML, non‐acute promyelocytic leukemia‐AML; WBC, white blood cells.
3.3. Association between the expression of PRR34‐AS1 and outcomes in AML patients
The median overall survival (OS) time for all AML patients was 10 months (range 1–90 months). Kaplan–Meier survival analysis showed that AML patients with high expression of PRR34‐AS1 had a significantly shorter OS than those with low expression of PRR34‐AS1 (p = 0.002; Figure 1B). To further explore the effect of PRR34‐AS1 expression on OS in AML patients, data from Gene Expression Omnibus (GEO; accession number GSE68833) were analyzed using GenomicScape online tool and similar results were obtained (Figure 1C). Cox regression analysis showed that high expression of PRR34‐AS1 was not an independent risk factor for OS in whole‐AML patients (Table 4).
TABLE 4.
Univariate and multivariate analyses of prognostic factors for overall survival in whole‐AML patients
| Variables | whole‐AML (n=83) | |||
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||
| HR (95% CI) | p value | HR (95% CI) | p value | |
| WBC | 2.514 (1.380–4.582) | 0.003 | 2.218 (1.221–4.029) | 0.009 |
| Age | 2.754 (1.494–5.075) | 0.001 | 1.173 (0.545–2.528) | 0.683 |
| PRR34‐AS1expression | 2.447 (1.313–4.559) | 0.004 | 1.573 (0.821–3.017) | 0.172 |
| Karyotype risk | 2.054 (1.401–3.011) | <0.001 | 2.070 (1.351–3.170) | <0.001 |
| CEBPA mutation | 1.123 (0.389–3.247) | 0.830 | — | — |
| NPM1 mutation | 1.522 (0.579–3.999) | 0.394 | — | — |
| FLT3‐ITD mutation | 1.113 (0.426–2.912) | 0.827 | — | — |
| c‐KIT mutation | 1.241 (0.167–9.202) | 0.833 | — | — |
| DNMT3A mutation | 1.228 (0.371–4.070) | 0.737 | — | — |
Variables including age (≤60 vs. <60 years), WBC (≥30 × 109 vs. <30 × 109/L), PRR34‐AS1 expression (low vs. high), karyotype risk (favorable vs. intermediate vs. poor), and gene mutations (mutant vs. wild type).
Multivariate analysis includes variables with p < 0.200 in univariate analysis.
Abbreviations: CI, confidence interval; HR, hazard ratio; WBC, white blood cells.
3.4. Association between the expression of PRR34‐AS1 and methylation of its promoter in AML
Analysis using GSE24006 and GSE63270 data sets showed high expression levels of PRR34‐AS1 in both data sets (Figure 1D,E). The methylation status of the PRR34‐AS1 promoter was determined to further explore whether changes in PRR34‐AS1 methylation affected its expression. MSP and BSP primer sets were designed and verified on the CpG island of the PRR34‐AS1 promoter region (Figure 2A). The methylation level of PRR34‐AS1 was then determined through MSP in 84 AML patients and 29 normal controls. Analysis showed that PRR34‐AS1 was hypomethylated in AML although there was no significant difference with normal control (p = 0.122; Figure 2B). Subsequently, two normal controls, two PRR34‐AS1‐hypermethylated AML patients, and two PRR34‐AS1‐unmethylated AML patients were randomly selected to verify the MSP results through BSP (Figure 2C). The unmethylated patients showed a completely unmethylated state in AML whereas hypermethylated patients and normal controls showed a higher density of methylation. Moreover, the degree of methylation in hypermethylated patients was lower compared with that in normal controls (Figure 2C). This implied that the results were consistent with MSP results. DiseaseMeth version 2.0 was used to determine the trend in the methylation of PRR34‐AS1 promoter (CpG island) in AML. The results revealed that AML patients had significantly lower PRR34‐AS1 methylation levels than the controls (Figure 2D). Furthermore, Spearman's rank test was used to analyze the correlation between the methylation and expression of PRR34‐AS1 in AML patients using the TCGA data sets. The findings showed that there was a significant negative correlation between the methylation and expression of PRR34‐AS1 (R = −0.236, p = 0.027, n = 168; Figure 2E). This finding implies that the aberrant methylation of PRR34‐AS1 may be an important mechanism for regulating its expression in AML.
FIGURE 2.
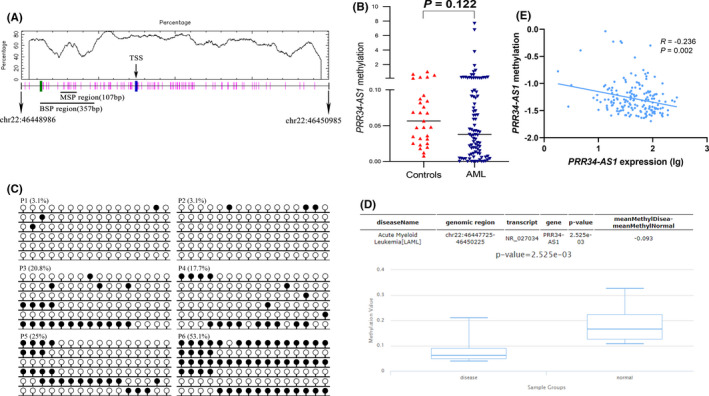
Validation of the methylation of PRR34‐AS1 in AML. (A) A schematic diagram of the CpG island in the promoter region of PRR34‐AS1. Vertical bars indicate CpG dinucleotides. Short horizontal lines represent corresponding positions amplified by MSP and BSP primers. The figure was generated using cpgplot (http://emboss.bioinformatics.nl/cgi‐bin/emboss/cpgplot) and Methyl Primer Express V1.0 software. TSS: transcription start site; MSP: methylation‐specific PCR; BSP: bisulfite sequencing PCR. B: Methylation levels of PRR34‐AS1 in the control group and AML patients were determined by MSP. (C) Methylation density of PRR34‐AS1 detected by BSP. The white cycle indicates unmethylated CpG dinucleotides whereas the black cycle represents methylated CpG dinucleotides P1 and P2: unmethylated AML patients; P3 and P4: methylated AML patients; P4 and P5: controls. (D) Methylation status of PRR34‐AS1 promoter (CpG island) was analyzed using Disease Meth version 2.0 tool (http://www.bio‐bigdata.com/diseasemeth/analyze.html). (E) Correlation analysis between PRR34‐AS1 gene expression and its methylation in AML patients was analyzed using data from the TCGA database. Spearman test was used for correlation analysis
3.5. Association between PRR34‐AS1 methylation and different clinical parameters in AML patients
The ROC curve was plotted to evaluate the diagnostic value of PRR34‐AS1 methylation in AML. The results showed that the PRR34‐AS1 methylation level may be a potential marker for distinguishing AML (especially non‐APL‐AML) patients from normal controls (95% CI = 0.513–0.722, p = 0.060, AUC = 0.617; 95% CI = 0.529–0.749, p = 0.032, AUC = 0.639; Figure 3A,B). Patients were then divided into PRR34‐AS1 hypermethylated group and hypomethylated group based on ROC analysis in order to explore the relationship between PRR34‐AS1 methylation and different clinical parameters in AML. Analysis showed no significant differences between the levels of methylation and gender, age, hemoglobin, platelets, and BM blasts between the two groups (p > 0.05; Table 5). Similarly, PRR34‐AS1 methylation showed no significant correlation with eight genetic mutations (p > 0.05; Table 5). However, patients with hypomethylated PRR34‐AS1 had a higher WBC count (p = 0.006) and showed a low frequency of favorable karyotypes compared with hypomethylated group [28% (7/25), p = 0.071; Table 5].
FIGURE 3.
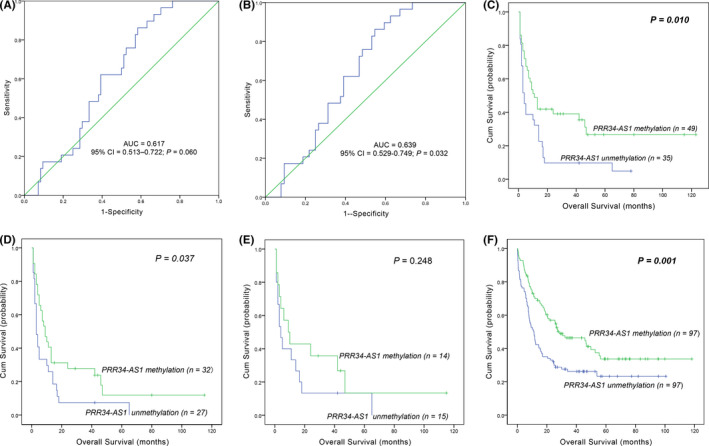
Effect of aberrant methylation of PRR34‐AS1 on the prognosis of AML. (A, B) A ROC curve of the clinical value of PRR34‐AS1 abnormal methylation in AML. (A) All AML patients; (B) non‐APL‐AML patients; AUC, area under the ROC curve; non‐APL, non‐acute promyelocytic leukemia; (C–E) effect of PRR34‐AS1 methylation on OS in AML. Patients were classified into PRR34‐AS1 hypomethylated and hypermethylated groups based on ROC curve analysis. (C) whole‐cohort AML patients. (D) non‐APL‐AML patients. (E) cytogenetically normal AML (CN‐AML) patients. (F) Effect of PRR34‐AS1 methylation on OS using TCGA database. One hundred and ninety‐four AML patients were grouped into hypomethylated and hypermethylated groups based on the median level of PRR34‐AS1 methylation, and survival analysis was conducted
TABLE 5.
Comparison of clinical characteristics between PRR34‐AS1 hypomethylated and PRR34‐AS1 hypermethylated group
| Patient's parameters | Hypermethylated (n = 49) | Hypomethylated (n = 35) | p value |
|---|---|---|---|
| Sex, male/female | 24/25 | 20/15 | 0.511 |
| Median hemoglobin, g/L (range) | 76 (32–138) | 78 (42–135) | 0.969 |
| Median age, years (range) | 52 (18–83) | 57 (20–85) | 0.162 |
| Median WBC, ×109/L (range) | 9.4 (0.3–107.0) | 35.6 (0.9–528) | 0.006 |
| Median platelets, ×109/L (range) | 34 (5–234) | 52 (9–264) | 0.211 |
| BM blasts, % (range) | 50 (1–94) | 35 (5.5–99.0) | 0.485 |
| FAB subtypes | 0.621 | ||
| M0 | 0 (0%) | 1 (2.9%) | |
| M1 | 4 (8.2%) | 2 (5.7%) | |
| M2 | 15 (30.6) | 15 (42.9%) | |
| M3 | 14 (28.6%) | 6 (17.1%) | |
| M4 | 11 (22.4%) | 6 (17.1%) | |
| M5 | 3 (6.1%) | 4 (11.4%) | |
| M6 | 2 (4.1%) | 1 (2.9%) | |
| Karyotype classification | 0.071 | ||
| Favorable | 18 (36.7%) | 7 (20%) | |
| Intermediate | 18 (36.7%) | 23 (65.7%) | |
| Poor | 10 (20.4%) | 4 (11.4%) | |
| No data | 3 (6.1%) | 1 (2.9%) | |
| Karyotype | 0.065 | ||
| Normal | 15 (30.6%) | 16 (45.7%) | |
| t (8;21) | 5 (10.2%) | 1 (2.9%) | |
| t (15;17) | 13 (26.5%) | 6 (17.1%) | |
| t (9;22) | 0 (0%) | 2 (5.7%) | |
| 11q23 | 1 (2%) | 0 (0%) | |
| −5/5q− | 1 (2%) | 0 (0%) | |
| −7/7q− | 3 (6.1%) | 1 (2.9%) | |
| complex | 2 (2.4%) | 6 (17.1%) | |
| No data | 9 (18.4%) | 0 (0%) | |
| Gene mutation | |||
| CEBPA (+/−) | 6/38 | 3/27 | 0.731 |
| NPM1 (+/−) | 2/42 | 2/28 | >0.999 |
| FLT3‐ITD (+/−) | 3/41 | 2/28 | >0.999 |
| C‐KIT (+/−) | 2/42 | 1/29 | >0.999 |
| N/K‐RAS (+/−) | 3/41 | 2/28 | >0.999 |
| IDH1/2 (+/−) | 5/44 | 2/28 | 0.161 |
| DNMT3A (+/−) | 2/37 | 1/24 | >0.999 |
| U2AF1 (+/−) | 1/38 | 2/24 | 0.562 |
| CR (+/−) | 18/21 | 9/17 | 0.154 |
Abbreviations: BM, bone marrow; CR, complete remission; FAB, French–American–British; WBC, white blood cells.
3.6. Association between PRR34‐AS1 methylation and clinical outcomes in AML patients
Correlation analysis was performed between PRR34‐AS1 methylation and clinical outcomes to explore the value of PRR34‐AS1 methylation in the prognosis of AML patients. Analysis methylation levels were not significantly correlated with CR of AML patients. Interestingly, OS of patients with hypomethylated PRR34‐AS1 was shorter than that of patients with hypermethylated PRR34‐AS1 in the whole‐cohort AML and non‐APL‐AML (p = 0.010, Figure 3C; p = 0.032; Figure 3D). Similar results were obtained through the analysis of the TCGA data sets (p < 0.001; Figure 3F). Moreover, Cox proportional hazards model supported that the hypomethylation of PRR34‐AS1 was an independent risk factor for OS among whole‐AML and non‐APL‐AML patients (Table 6).
TABLE 6.
Univariate and multivariate analyses of prognostic factors for overall survival in whole‐cohort‐AML and non‐APL patients
| Variables | Whole‐cohort‐AML (n = 84) | Non‐APL‐AML (n = 64) | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| HR (95% CI) | p value | HR (95% CI) | p value | HR (95% CI) | p value | HR (95% CI) | p value | |
| WBC | 1.895 (1.124–3.195) | 0.016 | 0.856 (0.443–1.652) | 0.643 | 1.443 (0.835–2.493) | 0.189 | 0.691 (0.341–1.402) | 0.306 |
| Age | 2.632 (1.550–4.470) | <0.001 | 1.846 (1.076–2.297) | 0.031 | 1.814 (1.042–3.158) | 0.035 | 1.370 (0.754–2.486) | 0.301 |
|
PRR34‐AS1 methylation |
0.522 (0.310–0.879) | 0.014 | 0.578 (0.329–1.017) | 0.057 | 0.573 (0.331–0.994) | 0.047 | 0.483 (0.264–0.883) | 0.018 |
| Karyotype risk | 1.754 (1.263–2.436) | 0.001 | 1.572 (1.076–2.297) | 0.019 | 1.496 (0.994–2.251) | 0.054 | 1.610 (1.009–2.570) | 0.046 |
| CEBPA mutation | 2.137 (0.896–5.096) | 0.087 | 1.828 (0.750–4.458) | 0.185 | 1.949 (0.811–4.685) | 0.136 | 2.139 (0.878–5.212) | 0.094 |
| NPM1 mutation | 1.453 (0.523–4.041) | 0.474 | — | — | 1.190 (0.422–3.354) | 0.743 | — | — |
| FLT3‐ITD mutation | 0.712 (0.256–1.980) | 0.515 | — | — | 0.833 (0.296–2.348) | 0.730 | — | — |
| c‐KIT mutation | 0.583 (0.142–2.405) | 0.456 | — | — | 0.388 (0.053–2.823) | 0.349 | — | — |
| N/K‐RAS mutation | 1.128 (0.403–3.152) | 0.819 | — | — | 0.918 (0.324–2.601) | 0.871 | ||
| DNMT3A mutation | 0.964 (0.346–2.682) | 0.944 | — | — | 0.770 (0.274–2.168) | 0.621 | — | — |
Variables including age (≤60 vs. <60 years), WBC (≥30×109 vs.<30×109/L), PRR34‐AS1 methylation (unmethylated vs. methylated), karyotype risk (favorable vs. intermediate vs. poor), and gene mutations (mutant vs. wild type).
Multivariate analysis includes variables with p < 0.200 in univariate analysis.
Abbreviations: AML, acute myeloid leukemia; CI, confidence interval; CN‐AML, cytogenetically normal AML; HR, hazard ratio; non‐APL‐AML, non‐acute promyelocytic leukemia‐AML; WBC, white blood cells.
4. DISCUSSION
AML is a complex disease with high heterogeneity at the molecular level and in clinical symptoms. 40 Most of the previous studies largely focused on protein‐coding genes as key components of disease progression. A few studies have explored the role of non‐coding genes in the progression of AML. Studies report that lncRNAs have a diagnostic value and prognostic potential in several types of cancer, including AML. 41
The present study explored the correlation between PRR34‐AS1 expression and prognosis of AML. Kaplan–Meier analysis showed that OS in AML patients with higher PRR34‐AS1 transcript level was significantly shorter compared with that of patients with low expression levels. Notably, high heterogeneity of AML may interfere with effective diagnosis, prognosis, and identification of predictive biomarkers. 42 Analysis of GEO and TCGA data sets showed a significant increase in the expression of PRR34‐AS1 in BM specimens of AML patients. In addition, patients with high expression of PRR34‐AS1 had a significantly shorter OS than the low expression group. However, Cox analysis showed that PRR34‐AS1 expression was not an independent factor affecting OS in AML patients. This finding implies that multiple molecular mechanisms may contribute to the differential expression of PRR34‐AS1, and PRR34‐AS1 may be involved in the early stages of AML disease progression. A previous study by Kang et al. used array comparative genomic hybridization to explore copy number variation (CNV) in PRR34‐AS1. The findings for the study showed that an increased copy number of PRR34‐AS1 was correlated with early recurrence and poor DFS in cholangiocarcinoma patients. 27 Findings of the present study showed that high expression of PRR34‐AS1 was associated with a reduced CR rate. Multivariate analysis further showed a significant correlation between the high expression of PRR34‐AS1 and low CR in AML patients. This suggested that high PRR34‐AS1 expression may be one of the related factors contributing to the poor efficacy of chemotherapy in AML patients. These findings demonstrated that high PRR34‐AS1 expression may be associated with poor chemotherapeutic efficacy and poor prognosis in AML patients. Notably, minimal residual disease (MRD) monitoring helps evaluate the efficacy of induction therapy and for monitoring the early recurrence of AML, to allow the adjustment of treatment strategies. However, there were fewer patients with serial samples in this study, the role of PRR34‐AS1 expression in MRD monitoring and recurrence of AML was not explored. Further studies with a longer follow‐up and a bigger sample size should explore the role of PRR34‐AS1 expression in MRD monitoring and recurrence of AML.
Previous studies report that epigenetic disorders play a vital role in the pathogenesis of AML. DNA methylation can be used as an epigenetic modification to regulate gene expression. 43 In this study, MSP and BSP were used to detect and verify the levels of methylation in the DMR of PRR34‐AS1. The relationship between the methylation levels of PRR34‐AS1 and the expression of this gene was also explored. Analysis showed that the DMR of PRR34‐AS1 displayed a pattern of hypomethylation, compared with the controls; however, there was no significant difference between the two groups. Spearman correlation analysis showed that PRR34‐AS1 hypomethylation was associated with its expression. This finding implies that hypomethylated DMR of PRR34‐AS1 may be an important regulatory mechanism for the expression of PRR34‐AS1 in AML. The effect of the abnormal methylation of PRR34‐AS1 on the prognosis of AML was explored. The findings showed that the hypomethylation of PRR34‐AS1 was correlated with a shorter OS of AML patients. Furthermore, multivariate analysis verified that PRR34‐AS1 hypomethylation was an independent risk factor for OS. However, the small sample size used in the study may have resulted in relative errors in the results. These results should, therefore, be verified using larger sample sizes. Similar results were obtained from analysis using DiseaseMeth version 2.0 and TCGA database. The findings showed a significant decrease in the methylation level of PRR34‐AS1 promoter in AML and the short OS for patients with hypomethylated PRR34‐AS1 compared with hypermethylated patients.
Two major hypomethylating agents (HMAs), decitabine and azacytidine, have been used clinically for the treatment of elderly AML patients not suitable for or decline intensive remission therapy. However, primary or secondary failure occurs in about 80% of treated patients. 44 Although reactivated tumor suppressor genes have been supposed as the major antileukemic mechanism of HMAs, there is a concern that specific oncogenes will also be reactivated by demethylation. 45 Preliminary findings of the current study show that PRR34‐AS1 may be an oncogenic lncRNAs. However, the exact role of PRR34‐AS1 in leukemogenesis should be explored further. Moreover, further studies should explore whether PRR34‐AS1 can be reactivated after treatment with HMA and the impact of its reactivation.
Although the present study uncovered some insightful findings, it had a number of shortcomings. First, the clinical sample size was small included patients with normal karyotypes and related gene mutations. Second, RQ‐PCR and other detection methods used in the study are less accurate than high‐throughput sequencing. Additionally, the experimental results were not verified through cell function experiments. Moreover, this was a preliminary study on the relationship between PRR34‐AS1 and AML. Therefore, more studies should be conducted to verify the results and uncover the underlying mechanisms of PRR34‐AS1.
In summary, the findings of this study show that high PRR34‐AS1 expression may be associated with poor chemotherapeutic efficacy and poor prognosis in AML patients. In addition, higher expression of PRR34‐AS1 was associated with the hypomethylation of its promoter, and hypomethylation of PRR34‐AS1 may affect the prognosis of AML patients.
DATA AVAILABILITY STATEMENT
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
J Q and F‐y N conceived and designed the experiments; F‐y N and YG performed the experiments; F‐y N, G‐k S, and Z‐j X analyzed the data; Z‐j X, J‐d Z, T‐j Z, and J‐y L collected the clinical data; J L, J‐c M, and J Q offered technique and language support; F‐y N wrote the paper. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Clinical Research Ethics Committee of the Affiliated People's Hospital of Jiangsu University.
CONSENT FOR PUBLICATION
Written informed consent was obtained from all enrolled individuals before their participation.
Supporting information
Table S1
ACKNOWLEDGMENTS
The authors would like to thank all the patients and their families for participating in this project.
Nan F‐Y, Gu Y, Xu Z‐J, et al. Abnormal expression and methylation of PRR34‐AS1 are associated with adverse outcomes in acute myeloid leukemia. Cancer Med. 2021;10:5283–5296. 10.1002/cam4.4085
Fang‐yu Nan, Yu Gu, Zi‐jun Xu are Contributed equally.
Funding information
This study was supported by the National Natural Science Foundation of China (81970118, 81900166, and 81900163), Medical Innovation Team of Jiangsu Province (CXTDB2017002), Zhenjiang Clinical Research Center of Hematology(SS2018009), Social Development Foundation of Zhenjiang (SH2019065,SH2019067), Scientific Research Project of The Fifth 169 Project of Zhenjiang (21).
Contributor Information
Jiang Lin, Email: qianjun0007@hotmail.com, Email: 2651329493@qq.com.
Jun Qian, Email: qianjun0007@hotmail.com.
REFERENCES
- 1. Khwaja A, Bjorkholm M, Gale RE. Acute myeloid leukaemia. Nat Rev Dis Primers. 2016;2(1). 10.1038/nrdp.2016.10. [DOI] [PubMed] [Google Scholar]
- 2. Dores GM, Devesa SS, Curtis RE, et al. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood. 2012;119(1):34‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112(6):2183‐2189. [DOI] [PubMed] [Google Scholar]
- 4. Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18(2):115‐136. [DOI] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research Network . Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059‐2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gourvest M, Brousset P, Bousquet M. Long noncoding RNAs in acute myeloid leukemia: functional characterization and clinical relevance. Cancers (Basel). 2019;11(11):1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsai C‐H, Yao C‐Y, Tien F‐M, et al. Incorporation of long non‐coding RNA expression profile in the 2017 ELN risk classification can improve prognostic prediction of acute myeloid leukemia patients. EBioMedicine. 2019;40:240‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the world health organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391‐2405. [DOI] [PubMed] [Google Scholar]
- 9. Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li CH, Chen Y. Insight into the role of long noncoding RNA in cancer development and progression. Int Rev Cell Mol Biol. 2016;326:33‐65. [DOI] [PubMed] [Google Scholar]
- 11. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965‐3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang TJ, Zhou JD, Zhang W, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou J, Xu J, Zhang L, et al. Combined single‐cell profiling of lncRNAs and functional screening reveals that H19 is pivotal for embryonic hematopoietic stem cell development. Cell Stem Cell. 2019;24(2):285‐298 e5. [DOI] [PubMed] [Google Scholar]
- 14. Wang SL, Huang Y, Su R, et al. Silencing long non‐coding RNA HOTAIR exerts anti‐oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int. 2019;19:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu S, Zheng C, Chen S, et al. Overexpression of long non‐coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol Lett. 2015;10(4):2410‐2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y‐Y, Huang S‐H, Zhou H‐R, et al. Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncol Rep. 2016;36(6):3113‐3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El‐Khazragy N, Elayat W, Matbouly S, et al. The prognostic significance of the long non‐coding RNAs "CCAT1, PVT1" in t(8;21) associated Acute Myeloid Leukemia. Gene. 2019;707:172‐177. [DOI] [PubMed] [Google Scholar]
- 18. Zeng C, Yu X, Lai J, et al. Overexpression of the long non‐coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng C, Xu Y, Xu L, et al. Inhibition of long non‐coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao C, Wang S, Zhao Y, et al. Long noncoding RNA NEAT1 modulates cell proliferation and apoptosis by regulating miR‐23a‐3p/SMC1A in acute myeloid leukemia. J Cell Physiol. 2019;234(5):6161‐6172. [DOI] [PubMed] [Google Scholar]
- 21. Pashaiefar H, Izadifard M, Yaghmaie M, et al. Low expression of long noncoding RNA IRAIN is associated with poor prognosis in non‐M3 acute myeloid leukemia patients. Genet Test Mol Biomarkers. 2018;22(5):288‐294. [DOI] [PubMed] [Google Scholar]
- 22. Yao H, Duan M, Lin L, et al. TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget. 2017;8(11):18337‐18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benetatos L, Hatzimichael E, Dasoula A, et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34(2):148‐153. [DOI] [PubMed] [Google Scholar]
- 24. Sellers ZP, Bolkun L, Kloczko J, et al. Increased methylation upstream of the MEG3 promotor is observed in acute myeloid leukemia patients with better overall survival. Clin Epigenetics. 2019;11(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP‐AS1 promotes the growth of hepatocellular carcinoma by targeting miR‐194‐5p/FOXA1 axis. Mol Cancer. 2019;18(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kesherwani V, Shukla M, Coulter DW, et al. Long non‐coding RNA profiling of pediatric Medulloblastoma. BMC Med Genomics. 2020;13(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang MJ, Kim J, Jang J‐Y, et al. 22q11‐q13 as a hot spot for prediction of disease‐free survival in bile duct cancer: integrative analysis of copy number variations. Cancer Genet. 2014;207(3):57‐69. [DOI] [PubMed] [Google Scholar]
- 28. Fang H, Zhang F‐X, Li H‐F, et al. PRR34‐AS1 overexpression promotes protection of propofol pretreatment against ischemia/reperfusion injury in a mouse model after total knee arthroplasty via blockade of the JAK1‐dependent JAK‐STAT signaling pathway. J Cell Physiol. 2020;235(3):2545‐2556. [DOI] [PubMed] [Google Scholar]
- 29. Cecotka A, Polanska J. Region‐specific methylation profiling in acute myeloid leukemia. Interdiscip Sci. 2018;10(1):33‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang ZH, Zhang W, Zhou JD, et al. Decreased SCIN expression, associated with promoter methylation, is a valuable predictor for prognosis in acute myeloid leukemia. Mol Carcinog. 2018;57(6):735‐744. [DOI] [PubMed] [Google Scholar]
- 31. Zhou LY, Zhai LL, Yin JY, et al. Pseudogene BMI1P1 expression as a novel predictor for acute myeloid leukemia development and prognosis. Oncotarget. 2016;7(30):47376‐47386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shafi A, Mitrea C, Nguyen T, et al. A survey of the approaches for identifying differential methylation using bisulfite sequencing data. Brief Bioinform. 2018;19(5):737‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qian J, Yao D‐M, Lin J, et al. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2012;7(9):e45760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin J, Yao D‐M, Qian J, et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6(10):e26906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang X, Qian J, Sun A, et al. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46(7–8):579‐583. [DOI] [PubMed] [Google Scholar]
- 36. Lin J, Yao D‐M, Qian J, et al. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91(4):519‐525. [DOI] [PubMed] [Google Scholar]
- 37. Wen XM, Lin J, Yang J, et al. Double CEBPA mutations are prognostically favorable in non‐M3 acute myeloid leukemia patients with wild‐type NPM1 and FLT3‐ITD. Int J Clin Exp Pathol. 2014;7(10):6832‐6840. [PMC free article] [PubMed] [Google Scholar]
- 38. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Medinger M, Passweg JR. Acute myeloid leukaemia genomics. Br J Haematol. 2017;179(4):530‐542. [DOI] [PubMed] [Google Scholar]
- 41. Mer AS, Lindberg J, Nilsson C, et al. Expression levels of long non‐coding RNAs are prognostic for AML outcome. J Hematol Oncol. 2018;11(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J, Lenferink AEG, Deng Y, et al. Corrigendum: Identification of high‐quality cancer prognostic markers and metastasis network modules. Nature. Communications. 2012;3(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang X, Wong MPM, Ng RK. Aberrant DNA methylation in acute myeloid leukemia and its clinical implications. Int J Mol Sci. 2019;20(18):4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nanah R, McCullough K, Hogan W, et al. Outcome of elderly patients after failure to hypomethylating agents given as frontline therapy for acute myeloid leukemia: Single institution experience. Am J Hematol. 2017;92(9):866‐871. [DOI] [PubMed] [Google Scholar]
- 45. Wen X‐M, Zhang T‐J, Ma J‐C, et al. Establishment and molecular characterization of decitabine‐resistant K562 cells. J Cell Mol Med. 2019;23(5):3317‐3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


