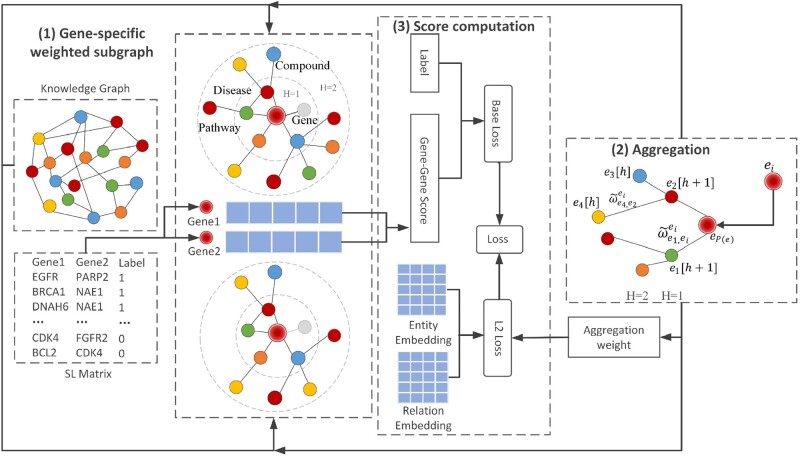
Fig. 1.
The framework of KG4SL. The workflow of KG4SL can be divided into three modules, including gene-specific weighted subgraph module, aggregation module and score computation module. (1) Gene-specific weighted subgraph: First, we construct a weighted subgraph from the KG. (2) Aggregation: Second, for each SL pair, we select the entities and relationships that are directly related to the nodes. Besides, we believe the biological information can flow between nodes through edges. Thus, we also aggregate the information of indirectly connected entities and relationships. Considering the problem of computing power, only two layers of entities and relationships are included. (3) Score computation: Third, the results of aggregation for two genes are used to compute their SL score through inner product. The loss function of KG4SL is composed of two kinds of losses, i.e. the base loss computed based on the truth label and the gene–gene score, and the L2 loss computed using the entity embedding, relation embedding and aggregation weights