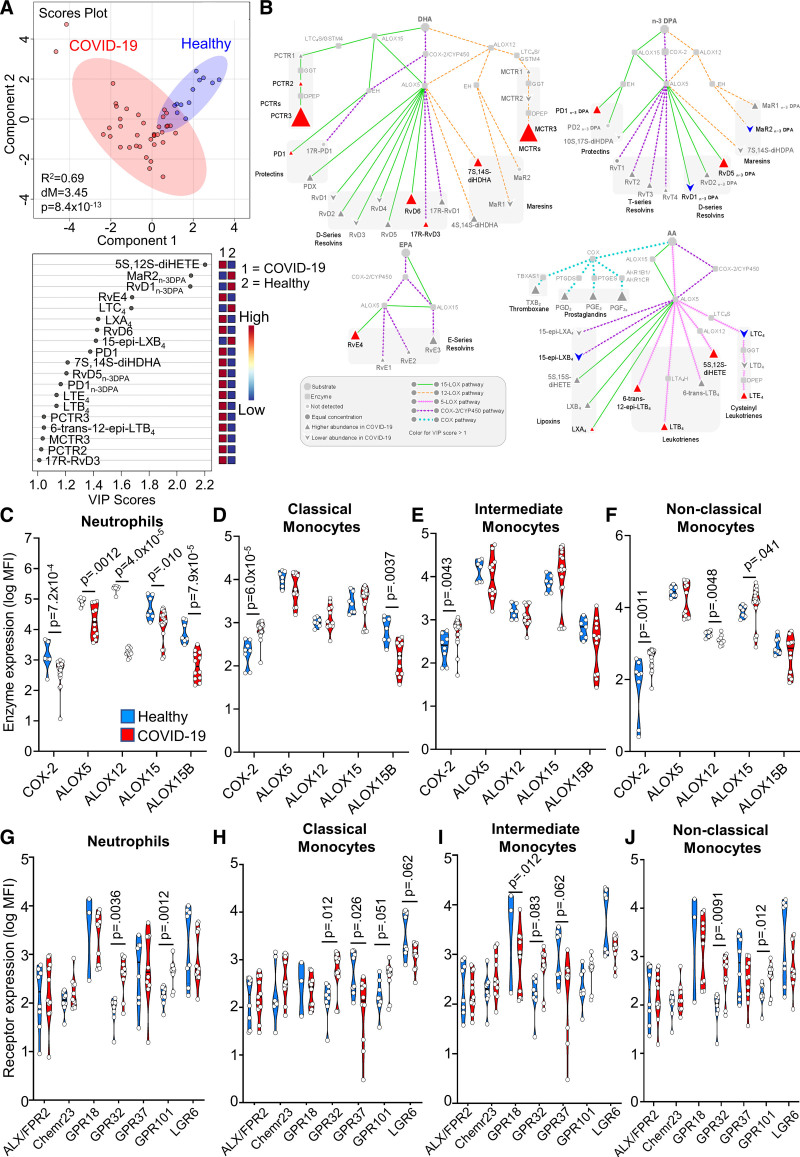
Figure 1.
Upregulation of specialized proresolving mediator (SPM) pathways in patients with coronavirus disease 2019 (COVID-19). Peripheral blood was collected from patients with COVID-19 or healthy volunteers and (A and B) plasma lipid mediators were identified and quantified using liquid chromatography tandem mass spectrometry-based mediator profiling (n=38 patients with COVID-19, n=12 healthy volunteers). A, Partial least squares discriminant analysis (PLS-DA) was performed on identified mediators. Top: Scores plot, shaded area represents 95% CI; R2, coefficient-of-determination; P, P value from Hotelling T2 test; dM, Mahalanobis distance between groups. Bottom: Variable Importance in Projection (VIP) scores plot. B, Analysis highlighting mediators with VIP scores >1 in PLS-DA and their biosynthetic pathways. C–J, Whole blood from healthy volunteers and patients with COVID-19 was incubated with lineage-marker antibodies against neutrophils (CD16+) and monocyte subsets (classical CD14++CD16+, intermediate CD14++CD16++, nonclassical CD14+CD16++) in combination with antibodies against (C–F) lipid mediator biosynthetic enzymes or (G–J) SPM receptors, and their expression was evaluated using flow cytometry. For (C–F), n=19 patients with COVID-19 (for COX-2, ALOX15), n=12 patients with COVID-19 (for ALOX5 [5-lipoxygenase], ALOX12, ALOX15B), and n=7 healthy volunteers. For (G–J), n=11 patients with COVID-19 (except for GPR18 and GPR37 where n=10) and n=8 healthy volunteers (except for GPR18 where n=3 healthy volunteers). Statistical differences between healthy and COVID-19 groups were established using Mann-Whitney test for each protein, and raw P values are displayed. GPCR indicates G protein-coupled receptor; MCTR, maresin conjugates in tissue regeneration; PCTR, protectin conjugates in tissue regeneration.