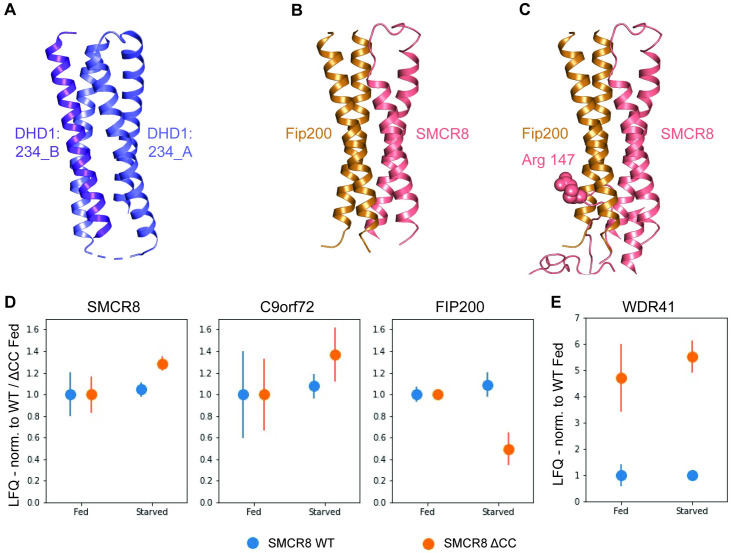
Fig 5. Structural and biochemical analysis of the SMCR8 coiled-coil region.
(A) Structure of an artificial 4 helix bundle generated by 2 monomers (PDB: 6DLC) (blue and purple) [39]. (B) Model of a 4-helix bundle formed by SMCR8N-C coiled coil (salmon pink) and FIP200 coiled-coil dimer (brown, PDB: 6GMA) [40] based on the artificial 4 helix bundle (PDB: 6DLC) [39]. (C) Model of SMCR8 coiled coil bound to FIP200 coiled-coil dimer along with the uDenn domain of SMCR8. Arg 147 of SMCR8 is shown in sphere representation. (D) Reduction in interaction between C9orf72-SMCR8ΔCC with FIP200 upon starvation. Fold change in the LFQ values of C9orf72 and SMCR8 Fl (WT, blue) or SMCR8ΔCC (ΔCC, orange) and FIP200 (normalized to the Fed condition) are shown. Standard error of the mean is shown. (E) Increased interaction between C9orf72-SMCR8ΔCC and WDR41. Fold change in the LFQ values of WDR41 (normalized to the WT Fed condition) is shown for both SMCR8 Fl (WT, blue) or SMCR8ΔCC (ΔCC, orange) pull-down experiments. Standard error of the mean is shown. Student t test: WDR41 Fed WT/ΔCC, p = 0.091; WDR41 Starved WT/ΔCC, p = 0.004; N = 3. The LFQ intensities and the corresponding analysis can be found in S2 Data. CC, coiled coil; Denn, differently expressed in normal and neoplastic cells; LFQ, label-free quantification; PDB, Protein Data Bank; uDenn, upstream Denn; WT, wild-type.