Fig. 3. Identification of GRB2 binding interfaces on MRE11, RPA70, and H2AX.
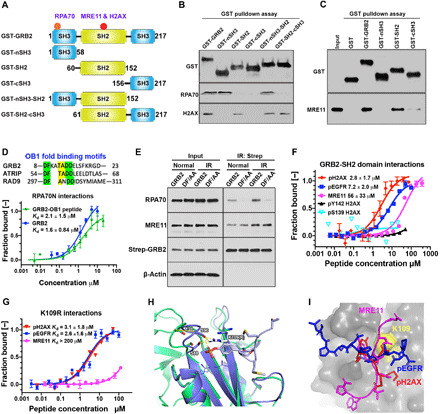
(A) Schematic DDR binding motifs (red circles) on GRB2 within its domain architecture. (B) GST-SH domains pulldown of HEK293T cell extracts followed by immunoblotting with indicated antibodies. (C) GST pulldown of bacterially purified human MRE11. (D) OB1 fold binding motif sequence alignment (top). MST isotherms of 50 nM Atto488–labeled RPA70 (residues 1 to 120) with titrating concentrations of GRB2 or GRB2-derived OB1 peptide. (E) Strep-Tactin precipitation of WT and DF/AA mutant from HEK293T cells that were either untreated (normal) or immediately after IR treated (5 Gy) and lysed, followed by Western blot detection with the indicated antibodies. (F) MST-binding isotherms of an Atto488-labeled GRB2-SH2 domain (100 nM) binding to synthetic peptides derived from indicated proteins. (G) MST isotherms of K109R binding to synthetic peptides derived from the indicated proteins. (H) Molecular docking overlay model showing S88 and S90 movement ~4.7 Å when bound to phosphotyrosine peptide. PDB structures used were as follows: 3wa4, purple; 1gri, green. K109R shown modeled (white sticks), which could engage with the loop containing S88 and S90 residues (purple sticks). (I) Docking of pH2AX (red), MRE11 (magenta), and pEGFR (blue) peptide into GRB2 SH2 domain pocket (gray). The relative docking orientation of each peptide is shown. Yellow surface marks the K109 position.
