Fig. 1.
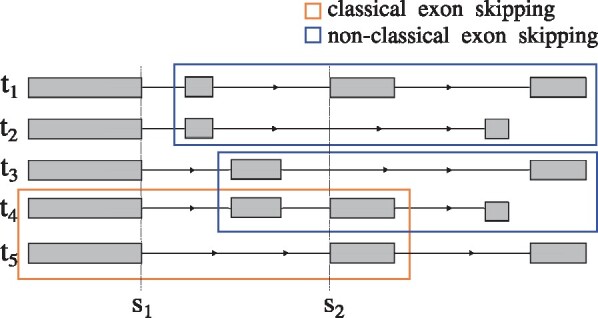
Complex alternative splicing involving five different transcripts. The two classical exon skipping events between t1 and t5, and between t4 and t5 do not fully capture the overall complexity. The two exon skippings marked in blue are not considered classical events and would not be reported by methods such as SplAdder, since they also differ in the last exon. Methods such as MAJIQ generalize simple events to more complex AS units that contain all introns sharing a common splice site. Two such AS units are required to describe the simple exon skipping event marked in orange, one comprising three introns sharing donor s1 and one containing three different introns sharing acceptor s2
