Fig. 5.
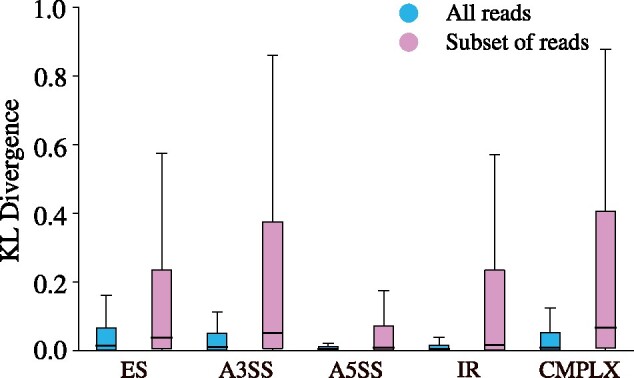
McSplicer leverages all RNA-seq reads mapped to a gene to improve the accuracy of splice site usage estimates. On the dataset with 50 million simulated reads, McSplicer achieves lower KL divergence from true splice site usages when considering all reads mapped to a gene locus at once (blue) compared to using only reads that overlap any of the event’s exons (pink). ES denotes exon skipping, A3SS alternative 3′ splice site, A5SS alternative 5′ splice site, IR intron retention and CMPLX complex events
