Figure 1. Carm1 is an epigenetic inhibitor in tumor-specific T cells.
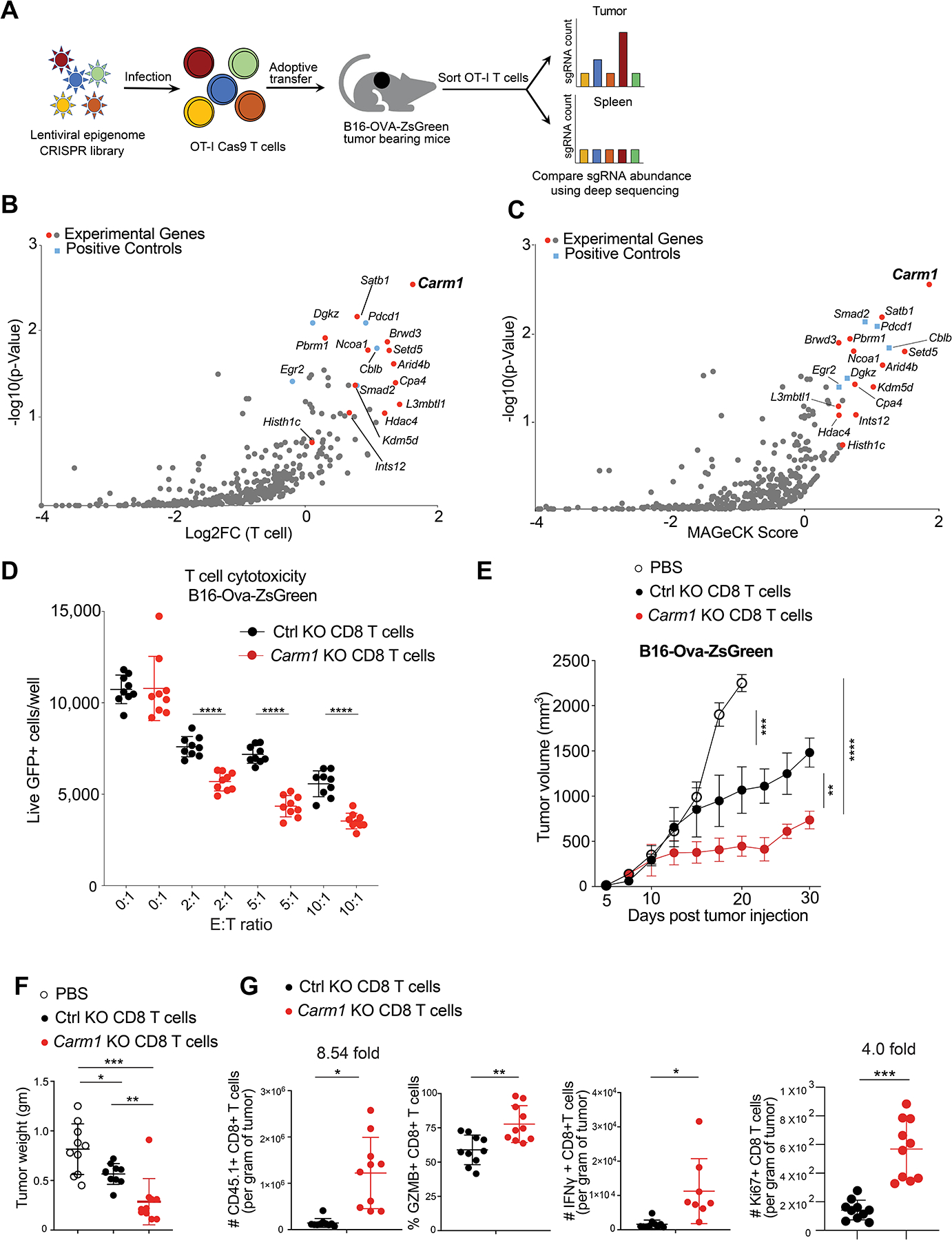
A. Experimental design for in vivo discovery of epigenetic regulators that inhibit CD8 T cell accumulation in tumors.
B. In vivo CRISPR screen with epigenetic gRNA library in tumor specific CD8 T cells. gRNA quantification in CD8 T cells was compared in tumors (experimental site) and spleens (control organ) (log2 fold change). Major experimental genes and positive control genes were highlighted in red and blue, respectively.
C. MAGeCK (Model-based Analysis of Genome-Wide CRIPSR-Cas9 knockout) analysis of in vivo CRISPR screen data; MAGeCK score provides integrated readout for strength of gene effects.
D. T cell cytotoxicity assay with Carm1-KO and control-KO OT-I CD8 T cells. CD8 T cells were edited by electroporation with Cas9 protein and bound gRNA, and cells were grown in IL-15 + IL-7 containing T cell media for 5 days. T cells were co-cultured with B16F10-OVA-ZsGreen tumor cells at indicated effector to target (E: T) ratios (n=8–10/replicates per condition); 24 hours later live GFP-positive tumor cells were counted using a Celigo image cytometer. Data are representative of three experiments and shown as mean ± SEM. ****p < 0.0001, by unpaired two-sided Mann-Whitney test.
E. Anti-tumor activity of adoptively transferred Carm1-KO or control-KO OT-I CD45.1 CD8 T cells. B16-OVA-ZsGreen tumor cells (0.1×106) were implanted subcutaneously. On day 7 post tumor cell inoculation, edited CD8 T cells (1×106) were transferred via tail vein injection. Tumor size was recorded; n=8–10 mice per group.
F. Tumor weights 7 days following adoptive T cell transfer for experiment shown in E.
G. Flow cytometry analysis of tumor-infiltrating Carm1-KO or control-KO CD8 T cells following adoptive transfer of edited OT-I CD45.1 CD8 T cells (n=10 mice/group) with gating on CD45.1 and CD8 T cell markers. Quantification of CD8 T cell infiltration and expression of effector (granzyme B, IFNγ) as well as proliferation (Ki-67) markers
