Figure 4. Innate immune activation in Carm1 deficient tumor cells.
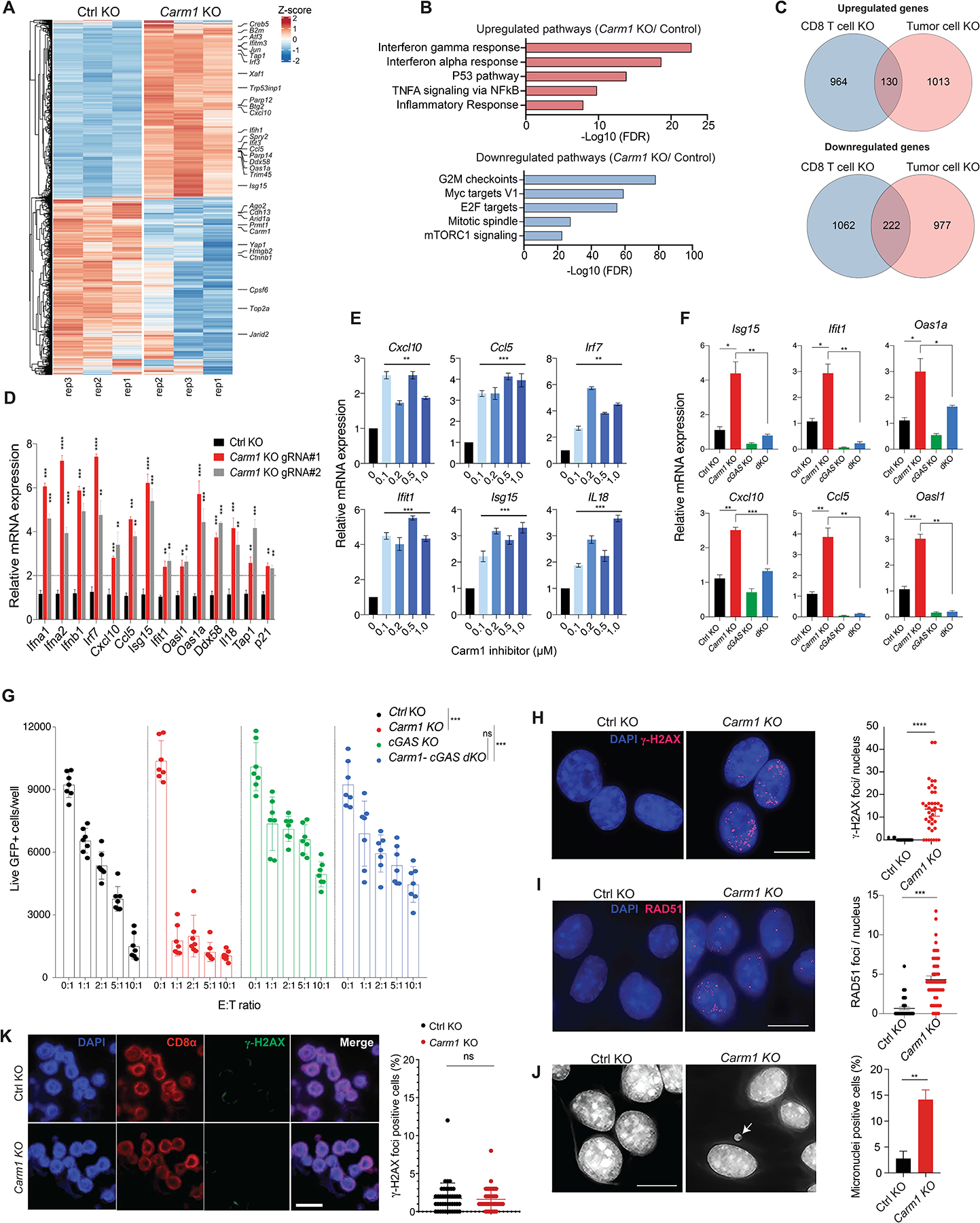
A. RNA-seq analysis of differentially expressed genes in Carm1-KO and control-KO B16F10 tumor cells (n=3/group). Data are representative of two independent experiments.
B. Gene Ontology (GO) analysis of significantly upregulated/downregulated genes in Carm1-KO compared to control-KO B16F10 tumor cells.
C. Venn diagram representing number of overlapping differentially expressed genes in Carm1-KO tumor and CD8 T cells.
D. Validation of interferon inducible genes (ISGs) by RT-qPCR in Carm1-KO compared to control-KO B16F10 cells (n=3/group).
E. RT-qPCR analysis of ISG mRNA levels following treatment of B16F10 cells with CARM1 inhibitor EZM2302 (0.1 – 1μM) or solvent control for 7 days (n=3/group).
F. Expression of selected ISGs in control-KO, Carm1-KO, cGAS-KO and Carm1/cGAS double-KO (dKO) B16F10 cells analyzed by RT-qPCR (n=3/group).
G. T cell cytotoxicity assay with control-KO, Carm1-KO, cGAS-KO and Carm1/cGAS dKO B16F10 cells. Tumor cells were co-cultured with OT-I CD8 T cells at indicated E:T ratios for 24 hours (n=7–10 replicates/condition); live GFP positive tumor cells were counted using a Celigo image cytometer. Data are shown as mean ± SEM. ***p < 0.001, by unpaired two-sided Mann-Whitney test.
H-I. dsDNA damage in Carm1-KO versus control-KO B16F10 tumor cells based on labeling with γH2AX (H) and RAD51 (I) Abs. Representative immunofluorescence images (left) of γH2AX or RAD51 antibody labeling (red); nuclei labeled with DAPI. Quantification of number of γH2AX or RAD51 foci/nucleus (right). Data are shown as mean ± SEM, ***p < 0.001, by unpaired two-sided Mann-Whitney test. Scale bar – 10μM.
J. Detection of micronuclei in Carm1-KO and control-KO B16F10 tumor cells. DNA was labeled with DAPI; representative images (left) and quantification of cells positive for micronuclei (right). Data are shown as mean ± SEM, **p < 0.01, by unpaired two-sided Mann-Whitney test. Scale bar – 10μM.
K. Analysis of Carm1-KO versus control-KO CD8+ T cells for dsDNA damage. OT-I CD8 T cells were plated 7 days posted editing using CRIPSR-Cas9 as previously described and stained for CD8α, γH2AX and DAPI. Representative immunofluorescence images of CD8α antibody labeling (red), γH2AX antibody labeling (green); nuclei stained with DAPI (blue). Scale bar – 20μM.
Data shown in D-J are representative of three independent experiments. Bar graphs represent data summarized as mean ± S.E.M. and were analyzed by unpaired two-sided Mann-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns (non-significant). Error bars for all qPCR data represent SD with three replicates per group.
