Figure 7. Relevance of CARM1 in human cancers.
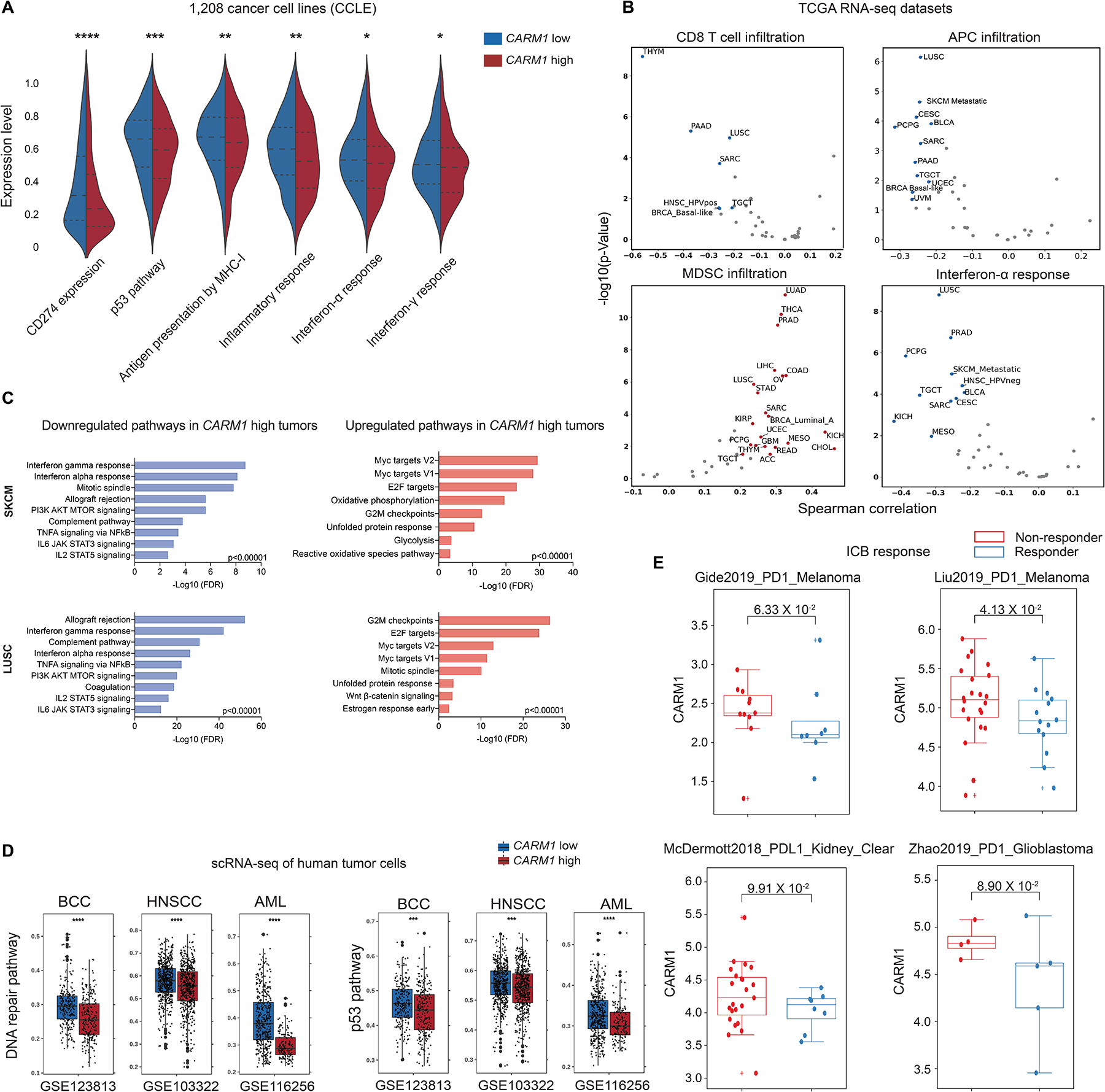
A. Analysis of indicated pathways across a diverse panel of 1,208 human cancer cell lines (Cancer Cell Line Encyclopedia, CCLE). Symmetric violin plots illustrate stratifications for CARM1 high and low cell lines using median expression levels.
B. Analysis of TCGA RNA-seq data across human cancer types. Correlation of CARM1 mRNA levels with indicated pathways. Plots show Spearman’s correlation and estimated statistical significance for indicated pathways in different cancer types adjusted for tumor purity. Each dot represents a cancer type in TCGA.
C. Gene Ontology (GO) analysis of significantly upregulated/downregulated genes in CARM1 high tumor cells in skin cutaneous melanoma (SKCM, N=442 patients) and lung squamous cell carcinoma (LUSC, N=363 patients) datasets (TCGA PanCancer Atlas).
D. Analysis of scRNA-seq data of malignant cells from three human cancer cohorts (GSE123813: basal cell carcinoma; GSE103322: head and neck cancer; GSE116256: AML). Scores for DNA repair, and p53 pathways are shown. Data were stratified by CARM1 high and low groups using median expression levels. Statistical comparisons were made using two-sided unpaired Mann-Whitney tests.
E. Correlation of CARM1 mRNA levels with response to ICB (immune checkpoint blockade) in cancer patients treated with PD-1 or PD-L1 mAbs. The analysis is shown for tumors with low (<median) MED12 mRNA levels. CARM1 mRNA levels did not correlate with response to ICB in tumors with high (>median) MED12 mRNA levels. The p-values were inferred by Mann-Whitney U test.
